7UXY
 
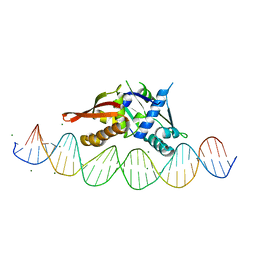 | | Isoreticular, interpenetrating co-crystal of protein variant Replication Initiator Protein REPE54 (L53G,Q54G,E55G) and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*P*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*P*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Snow, C.D. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
6RH6
 
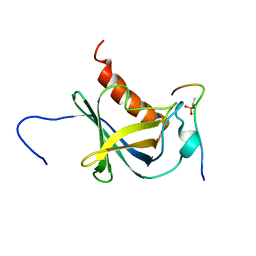 | | Solution structure and 1H, 13C and 15N chemical shift assignments for the complex of NECAP1 PHear domain with phosphorylated AP2 mu2 148-163 | | Descriptor: | AP-2 complex subunit mu, Adaptin ear-binding coat-associated protein 1 | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Herrmann, T. | | Deposit date: | 2019-04-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
6RH5
 
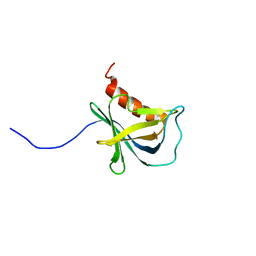 | | Solution structure and 1H, 13C and 15N chemical shift assignments for NECAP1 PHear domain | | Descriptor: | Adaptin ear-binding coat-associated protein 1 | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Herrmann, T. | | Deposit date: | 2019-04-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Temporal Ordering in Endocytic Clathrin-Coated Vesicle Formation via AP2 Phosphorylation.
Dev.Cell, 50, 2019
|
|
7ZFM
 
 | | Engineered Protein Targeting the Zika Viral Envelope Fusion Loop | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, HEXAETHYLENE GLYCOL, ... | | Authors: | Athayde, D, Archer, M, Viana, I.F.T, Adan, W.C.S, Xavier, L.S.S, Lins, R.D. | | Deposit date: | 2022-04-01 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | In Vitro Neutralisation of Zika Virus by an Engineered Protein Targeting the Viral Envelope Fusion Loop
SSRN, 2022
|
|
8AKM
 
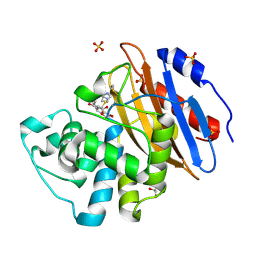 | | Acyl-enzyme complex of ertapenem bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, Ertapenem, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
8AKK
 
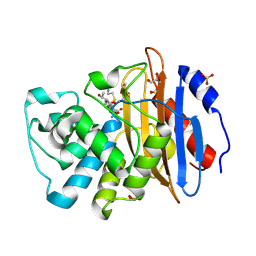 | | Acyl-enzyme complex of imipenem bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
8AKJ
 
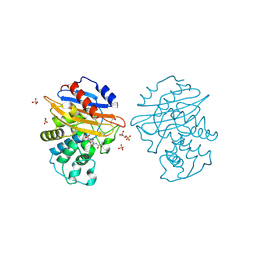 | | Acyl-enzyme complex of cephalothin bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | 5-METHYLENE-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-5,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
8AKL
 
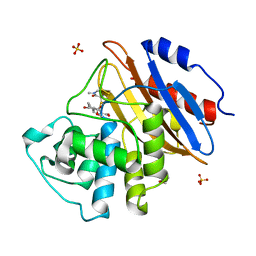 | | Acyl-enzyme complex of meropenem bound to deacylation mutant KPC-2 (E166Q) | | Descriptor: | (2S,3R,4R)-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl-3-methyl-2-[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Tautomer-Specific Deacylation and Omega-Loop Flexibility Explain the Carbapenem-Hydrolyzing Broad-Spectrum Activity of the KPC-2 beta-Lactamase.
J.Am.Chem.Soc., 145, 2023
|
|
7RGV
 
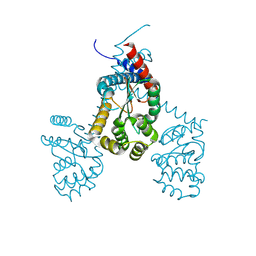 | |
4FE1
 
 | | Improving the Accuracy of Macromolecular Structure Refinement at 7 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Adams, P.D, Fromme, P, Levitt, M, Schroeder, G.F, Brunger, A.T. | | Deposit date: | 2012-05-29 | | Release date: | 2012-08-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.9228 Å) | | Cite: | Improving the accuracy of macromolecular structure refinement at 7 A resolution.
Structure, 20, 2012
|
|
9FHT
 
 | |
9FI4
 
 | |
9FI5
 
 | |
9QEM
 
 | | Crystal structure of YTHDF2 in complex with compound 3 (AI-DF2-11) | | Descriptor: | 5-[[(3-methoxyphenyl)amino]methylidene]-2-sulfanylidene-1,3-diazinane-4,6-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nai, F, Invernizzi, A, Caflisch, A. | | Deposit date: | 2025-03-10 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of YTHDF2 Ligands by Fragment-Based Design
Acs Bio Med Chem Au, 2025
|
|
9QEO
 
 | |
9QFL
 
 | | Crystal structure of YTHDF2 in complex with compound 15 (AI-DF2-68) | | Descriptor: | 6-sulfanylidene-1,7-dihydropyrazolo[3,4-d]pyrimidin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Nai, F, Invernizzi, A, Caflisch, A. | | Deposit date: | 2025-03-11 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of YTHDF2 Ligands by Fragment-Based Design
Acs Bio Med Chem Au, 2025
|
|
9DDR
 
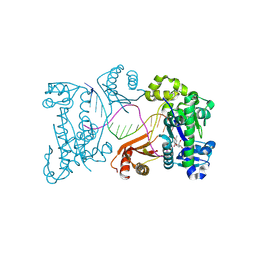 | | Ternary substrate complex of DNA polymerase iota with DNA (template A), Ca2+, and dTTP | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA polymerase iota, ... | | Authors: | Frevert, Z, Reusch, D, Freudenthal, B, Washington, M.T. | | Deposit date: | 2024-08-28 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Visualizing DNA polymerase iota catalyze Hoogsteen-directed DNA synthesis
Nat Commun, 16, 2025
|
|
9FHU
 
 | |
9FHW
 
 | |
9FHV
 
 | |
9FHX
 
 | |
9FHY
 
 | |
9FHZ
 
 | |
9FI0
 
 | |
9FI2
 
 | |
