1H7W
 
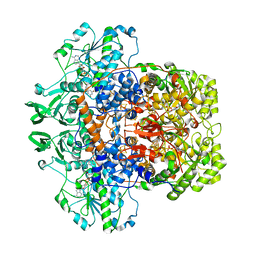 | | Dihydropyrimidine dehydrogenase (DPD) from pig | | Descriptor: | DIHYDROPYRIMIDINE DEHYDROGENASE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Schneider, G, Schnackerz, K.D, Lindqvist, Y. | | Deposit date: | 2001-01-19 | | Release date: | 2001-02-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil.
EMBO J., 20, 2001
|
|
1DAK
 
 | | DETHIOBIOTIN SYNTHETASE FROM ESCHERICHIA COLI, COMPLEX REACTION INTERMEDIATE ADP AND MIXED ANHYDRIDE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DETHIOBIOTIN SYNTHETASE, MAGNESIUM ION, ... | | Authors: | Kaeck, H, Gibson, K.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 1998-03-31 | | Release date: | 1999-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Snapshot of a phosphorylated substrate intermediate by kinetic crystallography.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2WG9
 
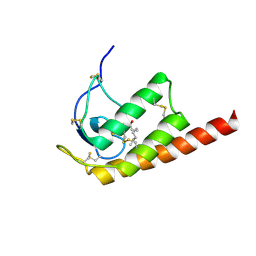 | | Structure of Oryza Sativa (Rice) PLA2, complex with octanoic acid | | Descriptor: | CALCIUM ION, OCTANOIC ACID (CAPRYLIC ACID), PUTATIVE PHOSPHOLIPASE A2, ... | | Authors: | Guy, J.E, Stahl, U, Lindqvist, Y. | | Deposit date: | 2009-04-16 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Class Xib Phospholipase A2 (Pla2): Rice (Oryza Sativa) Isoform-2 Pla2 and an Octanoate Complex.
J.Biol.Chem., 284, 2009
|
|
1GV9
 
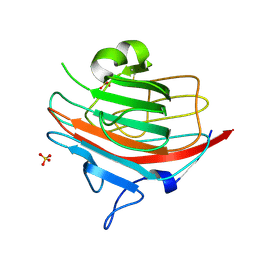 | | p58/ERGIC-53 | | Descriptor: | P58/ERGIC-53, SULFATE ION | | Authors: | Velloso, L.M, Svensson, K, Schneider, G, Pettersson, R.F, Lindqvist, Y. | | Deposit date: | 2002-02-07 | | Release date: | 2002-02-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Carbohydrate Recognition Domain of P58/Ergic-53, a Protein Involved in Glycoprotein Export from the Endoplasmic Reticulum.
J.Biol.Chem., 277, 2002
|
|
1E5M
 
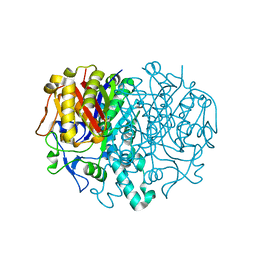 | |
1YBV
 
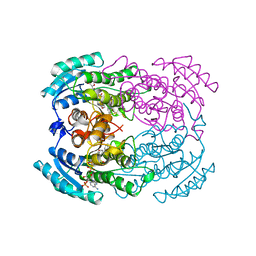 | | STRUCTURE OF TRIHYDROXYNAPHTHALENE REDUCTASE IN COMPLEX WITH NADPH AND AN ACTIVE SITE INHIBITOR | | Descriptor: | 5-METHYL-1,2,4-TRIAZOLO[3,4-B]BENZOTHIAZOLE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIHYDROXYNAPHTHALENE REDUCTASE | | Authors: | Andersson, A, Schneider, G, Lindqvist, Y. | | Deposit date: | 1996-09-23 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the ternary complex of 1,3,8-trihydroxynaphthalene reductase from Magnaporthe grisea with NADPH and an active-site inhibitor.
Structure, 4, 1996
|
|
1H75
 
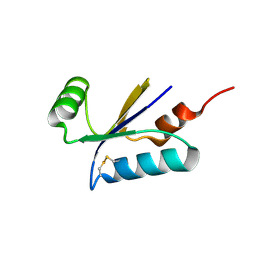 | | Structural basis for the thioredoxin-like activity profile of the glutaredoxin-like protein NrdH-redoxin from Escherichia coli. | | Descriptor: | GLUTAREDOXIN-LIKE PROTEIN NRDH | | Authors: | Stehr, M, Schneider, G, Aslund, F, Holmgren, A, Lindqvist, Y. | | Deposit date: | 2001-07-03 | | Release date: | 2001-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Thioredoxin-Like Activity Profile of the Glutaredoxin-Like Nrdh-Redoxin from Escherichia Coli
J.Biol.Chem., 276, 2001
|
|
1E5Q
 
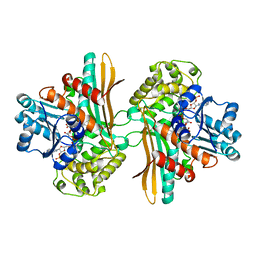 | | Ternary complex of saccharopine reductase from Magnaporthe grisea, NADPH and saccharopine | | Descriptor: | N-(5-AMINO-5-CARBOXYPENTYL)GLUTAMIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Saccharopine dehydrogenase [NADP(+), ... | | Authors: | Johansson, E, Steffens, J.J, Lindqvist, Y, Schneider, G. | | Deposit date: | 2000-07-28 | | Release date: | 2000-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Saccharopine Reductase from Magnaporthe Grisea, an Enzyme of the Alpha-Aminoadipate Pathway of Lysine Biosynthesis
Structure, 8, 2000
|
|
2W0U
 
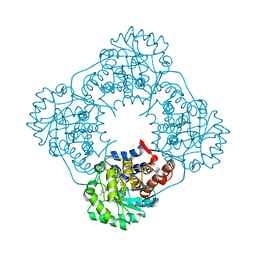 | | CRYSTAL STRUCTURE OF HUMAN GLYCOLATE OXIDASE IN COMPLEX WITH THE INHIBITOR 5-[(4-CHLOROPHENYL)SULFANYL]- 1,2,3-THIADIAZOLE-4-CARBOXYLATE. | | Descriptor: | 5-[(4-chlorophenyl)sulfanyl]-1,2,3-thiadiazole-4-carboxylate, FLAVIN MONONUCLEOTIDE, HYDROXYACID OXIDASE 1 | | Authors: | Bourhis, J.M, Lindqvist, Y. | | Deposit date: | 2008-10-10 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of Human Glycolate Oxidase in Complex with the Inhibitor 4-Carboxy-5-[(4-Chlorophenyl)Sulfanyl]-1,2,3-Thiadiazole.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2WG8
 
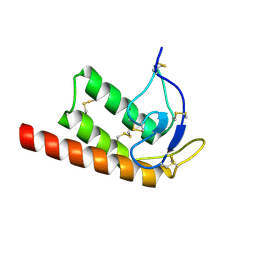 | | Structure of Oryza Sativa (Rice) PLA2, orthorhombic crystal form | | Descriptor: | CALCIUM ION, PUTATIVE PHOSPHOLIPASE A2, SODIUM ION | | Authors: | Guy, J.E, Stahl, U, Lindqvist, Y. | | Deposit date: | 2009-04-16 | | Release date: | 2009-06-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Class Xib Phospholipase A2 (Pla2): Rice (Oryza Sativa) Isoform-2 Pla2 and an Octanoate Complex.
J.Biol.Chem., 284, 2009
|
|
4STD
 
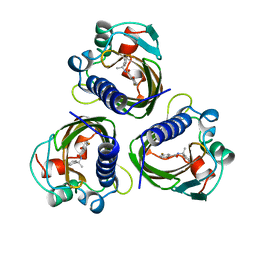 | | HIGH RESOLUTION STRUCTURES OF SCYTALONE DEHYDRATASE-INHIBITOR COMPLEXES CRYSTALLIZED AT PHYSIOLOGICAL PH | | Descriptor: | N-[1-(4-BROMOPHENYL)ETHYL]-5-FLUORO SALICYLAMIDE, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-10 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
2WG7
 
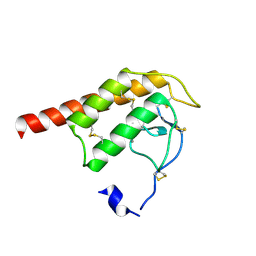 | | Structure of Oryza Sativa (Rice) PLA2 | | Descriptor: | CALCIUM ION, PUTATIVE PHOSPHOLIPASE A2, SODIUM ION | | Authors: | Guy, J.E, Stahl, U, Lindqvist, Y. | | Deposit date: | 2009-04-16 | | Release date: | 2009-06-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Class Xib Phospholipase A2 (Pla2): Rice (Oryza Sativa) Isoform-2 Pla2 and an Octanoate Complex.
J.Biol.Chem., 284, 2009
|
|
1CK7
 
 | | GELATINASE A (FULL-LENGTH) | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (GELATINASE A), ... | | Authors: | Morgunova, E, Tuuttila, A, Bergmann, U, Isupov, M, Lindqvist, Y, Schneider, G, Tryggvason, K. | | Deposit date: | 1999-04-28 | | Release date: | 1999-08-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human pro-matrix metalloproteinase-2: activation mechanism revealed.
Science, 284, 1999
|
|
2C31
 
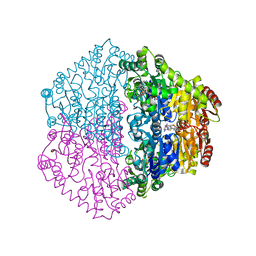 | | CRYSTAL STRUCTURE OF OXALYL-COA DECARBOXYLASE IN COMPLEX WITH THE COFACTOR DERIVATIVE THIAMIN-2-THIAZOLONE DIPHOSPHATE AND ADENOSINE DIPHOSPHATE | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Berthold, C.L, Moussatche, P, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2005-10-03 | | Release date: | 2005-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for Activation of the Thiamin Diphosphate-Dependent Enzyme Oxalyl-Coa Decarboxylase by Adenosine Diphosphate.
J.Biol.Chem., 280, 2005
|
|
1DD8
 
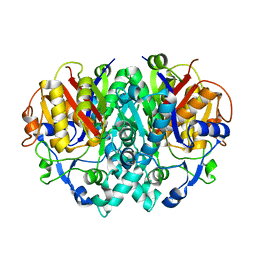 | | CRYSTAL STRUCTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I FROM ESCHERICHIA COLI | | Descriptor: | BETA-KETOACYL [ACYL CARRIER PROTEIN] SYNTHASE I | | Authors: | Olsen, J.G, Kadziola, A, von Wettstein-Knowles, P, Siggaard-Andersen, M, Lindquist, Y, Larsen, S. | | Deposit date: | 1999-11-09 | | Release date: | 1999-11-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray crystal structure of beta-ketoacyl [acyl carrier protein] synthase I.
FEBS Lett., 460, 1999
|
|
1KO1
 
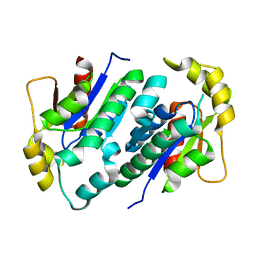 | | Crystal structure of gluconate kinase | | Descriptor: | CHLORIDE ION, Gluconate kinase | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1KOF
 
 | | Crystal structure of gluconate kinase | | Descriptor: | Gluconate kinase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1KO4
 
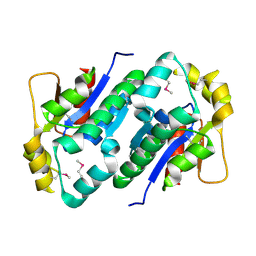 | | Crystal structure of gluconate kinase | | Descriptor: | CHLORIDE ION, Gluconate kinase | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
4HU2
 
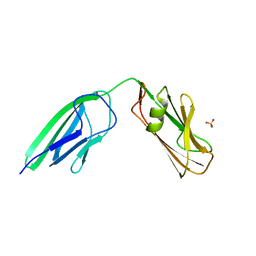 | | Crystal structure of LdtMt2, a L,D-transpeptidase from Mycobacterium tuberculosis: domain A and B | | Descriptor: | PROBABLE CONSERVED LIPOPROTEIN LPPS, SULFATE ION | | Authors: | Both, D, Steiner, E, Lindqvist, Y, Schnell, R, Schneider, G. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of LdtMt2, an L,D-transpeptidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1ZJ8
 
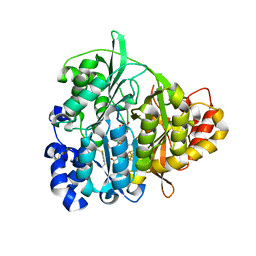 | | Structure of Mycobacterium tuberculosis NirA protein | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Probable ferredoxin-dependent nitrite reductase NirA, ... | | Authors: | Schnell, R, Sandalova, T, Hellman, U, Lindqvist, Y, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Siroheme- and [Fe4-S4]-dependent NirA from Mycobacterium tuberculosis Is a Sulfite Reductase with a Covalent Cys-Tyr Bond in the Active Site
J.Biol.Chem., 280, 2005
|
|
5STD
 
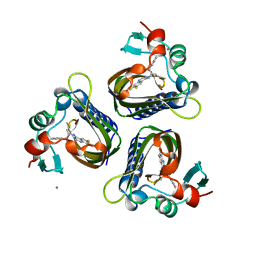 | | SCYTALONE DEHYDRATASE PLUS INHIBITOR 2 | | Descriptor: | (6,7-DIFLUORO-QUINAZOLIN-4-YL)-(1-METHYL-2,2-DIPHENYL-ETHYL)-AMINE, CALCIUM ION, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-10 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
2J2F
 
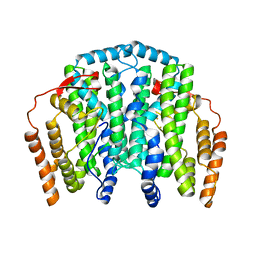 | | The T199D Mutant of Stearoyl Acyl Carrier Protein Desaturase from Ricinus Communis (Castor Bean) | | Descriptor: | ACYL-[ACYL-CARRIER-PROTEIN] DESATURASE, FE (III) ION | | Authors: | Guy, J.E, Abreu, I.A, Moche, M, Lindqvist, Y, Whittle, E, Shanklin, J. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Single Mutation in the Castor {Delta}9-18:0- Desaturase Changes Reaction Partitioning from Desaturation to Oxidase Chemistry.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1ZJ9
 
 | | Structure of Mycobacterium tuberculosis NirA protein | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Probable ferredoxin-dependent nitrite reductase NirA, ... | | Authors: | Schnell, R, Sandalova, T, Hellman, U, Lindqvist, Y, Schneider, G. | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Siroheme- and [Fe4-S4]-dependent NirA from Mycobacterium tuberculosis Is a Sulfite Reductase with a Covalent Cys-Tyr Bond in the Active Site
J.Biol.Chem., 280, 2005
|
|
4HUC
 
 | | Crystal structure of LdtMt2, a L,D-transpeptidase from Mycobacterium tuberculosis: domain B and C | | Descriptor: | ACETATE ION, PROBABLE CONSERVED LIPOPROTEIN LPPS, SODIUM ION | | Authors: | Both, D, Steiner, E, Lindqvist, Y, Schnell, R, Schneider, G. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of LdtMt2, an L,D-transpeptidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2JIB
 
 | | X-ray structure of Oxalyl-CoA decarboxylase in complex with coenzyme- A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COENZYME A, MAGNESIUM ION, ... | | Authors: | Berthold, C.L, Toyota, C.G, Moussatche, P, Wood, M.D, Leeper, F, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-02-26 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic Snapshots of Oxalyl-Coa Decarboxylase Give Insights Into Catalysis by Nonoxidative Thdp-Dependent Decarboxylases
Structure, 15, 2007
|
|
