3SY4
 
 | | Crystal structure of sulfide:quinone oxidoreductase Ser126Ala variant from Acidithiobacillus ferrooxidans | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-15 | | Release date: | 2012-05-16 | | Last modified: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3SZF
 
 | | Crystal structure of sulfide:quinone oxidoreductase H198A variant from Acidithiobacillus ferrooxidans in complex with bound trisulfide and decylubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-18 | | Release date: | 2012-05-16 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (2.0994 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3SZW
 
 | | Crystal structure of sulfide:quinone oxidoreductase Cys128Ser variant from Acidithiobacillus ferrooxidans in complex with decylubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-19 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3SYI
 
 | | Crystal structure of sulfide:quinone oxidoreductase Ser126Ala variant from Acidithiobacillus ferrooxidans using 7.0 keV diffraction data | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-17 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2001 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3T0K
 
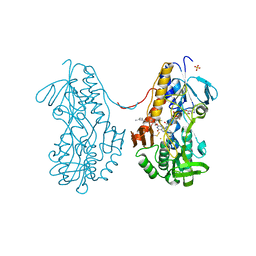 | | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans with bound trisulfide and decylubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, FLAVIN-ADENINE DINUCLEOTIDE, HYDROSULFURIC ACID, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-20 | | Release date: | 2012-05-16 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3SZ0
 
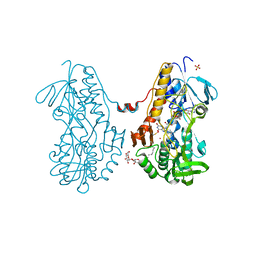 | | Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans in complex with sodium selenide | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, SELENIUM ATOM, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-18 | | Release date: | 2012-05-16 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.1501 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
3SX6
 
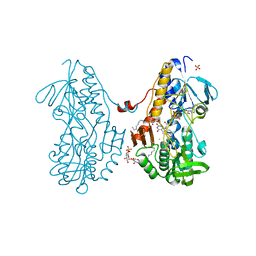 | | Crystal structure of sulfide:quinone oxidoreductase Cys356Ala variant from Acidithiobacillus ferrooxidans complexed with decylubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cherney, M.M, Zhang, Y, James, M.N.G, Weiner, J.H. | | Deposit date: | 2011-07-14 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7955 Å) | | Cite: | Structure-activity characterization of sulfide:quinone oxidoreductase variants.
J.Struct.Biol., 178, 2012
|
|
2H9H
 
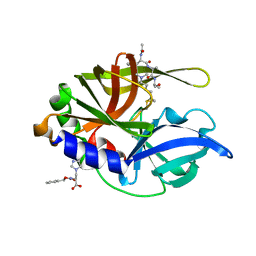 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | Hepatitis A virus protease 3C, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, Three residue peptide | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-06-09 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | An Episulfide Cation (Thiiranium Ring) Trapped in the Active Site of HAV 3C Proteinase Inactivated by Peptide-based Ketone Inhibitors.
J.Mol.Biol., 361, 2006
|
|
3EI9
 
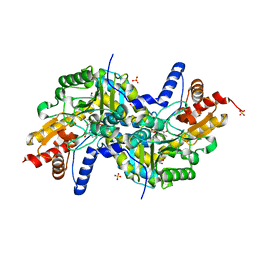 | | Crystal structure of K270N variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with L-Glu: External aldimine form | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-glutamic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EI8
 
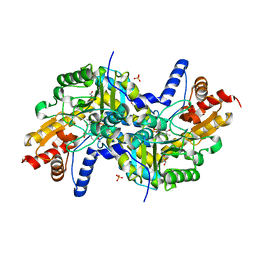 | | Crystal structure of K270N variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with LL-DAP: External aldimine form | | Descriptor: | (2S,6S)-2-amino-6-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}heptanedioic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EI5
 
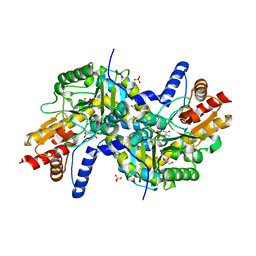 | | Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with PLP-Glu: an external aldimine mimic | | Descriptor: | GLYCEROL, LL-diaminopimelate aminotransferase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EIA
 
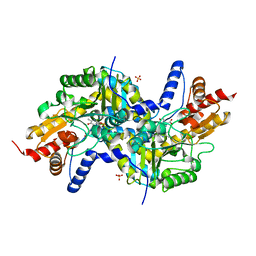 | | Crystal structure of K270Q variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with L-Glu: External aldimine form | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-glutamic acid, LL-diaminopimelate aminotransferase, SULFATE ION | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EI7
 
 | | Crystal structure of apo-LL-diaminopimelate aminotransferase from Arabidopsis thaliana (no PLP) | | Descriptor: | LL-diaminopimelate aminotransferase, SULFATE ION | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3CAG
 
 | | Crystal structure of the oligomerization domain hexamer of the arginine repressor protein from Mycobacterium tuberculosis in complex with 9 arginines. | | Descriptor: | ARGININE, Arginine repressor | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Lu, G.J, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-02-19 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of the arginine repressor protein from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3EJX
 
 | | Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana in complex with LL-AziDAP | | Descriptor: | (2S,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, Diaminopimelate epimerase, chloroplastic | | Authors: | Pillai, B, Moorthie, V.A, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana, an amino acid racemase critical for L-lysine biosynthesis.
J.Mol.Biol., 385, 2009
|
|
3EKM
 
 | | Crystal structure of diaminopimelate epimerase form arabidopsis thaliana in complex with irreversible inhibitor DL-AziDAP | | Descriptor: | (2R,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, Diaminopimelate epimerase, chloroplastic | | Authors: | Pillai, B, Moorthie, V.A, Cherney, M.M, van Belkum, M.J, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-19 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana, an amino acid racemase critical for L-lysine biosynthesis.
J.Mol.Biol., 385, 2009
|
|
3FHZ
 
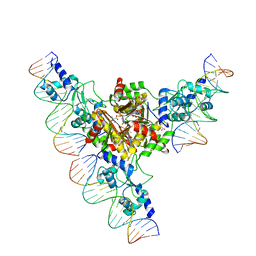 | | Crystal structure of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator and co-repressor, L-arginine | | Descriptor: | 5'-D(*TP*GP*TP*TP*GP*CP*AP*TP*AP*AP*CP*GP*AP*TP*GP*CP*AP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*CP*AP*TP*CP*GP*TP*TP*AP*TP*GP*CP*AP*AP*CP*A)-3', ACETATE ION, ... | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-12-10 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | The structure of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator and Co-repressor, L-arginine.
J.Mol.Biol., 388, 2009
|
|
3EI6
 
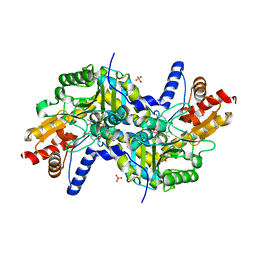 | | Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with PLP-DAP: an external aldimine mimic | | Descriptor: | (2S,6S)-2-amino-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]heptanedioic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EIB
 
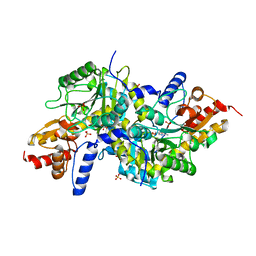 | | Crystal structure of K270N variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana | | Descriptor: | GLYCEROL, LL-diaminopimelate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
1HTR
 
 | |
1K2I
 
 | | Crystal Structure of Gamma-Chymotrypsin in Complex with 7-Hydroxycoumarin | | Descriptor: | 2,4-DIHYDROXY-TRANS CINNAMIC ACID, CHYMOTRYPSINOGEN A, SULFATE ION | | Authors: | Ghani, U, Ng, K.K.S, Atta-ur-Rahman, Choudhary, M.I, Ullah, N, James, M.N.G. | | Deposit date: | 2001-09-27 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of gamma-chymotrypsin in complex with 7-hydroxycoumarin.
J.Mol.Biol., 314, 2001
|
|
1L1N
 
 | | POLIOVIRUS 3C PROTEINASE | | Descriptor: | Genome polyprotein: Picornain 3C | | Authors: | Mosimann, S.C, Chernaia, M.M, Sia, S, Plotch, S, James, M.N.G. | | Deposit date: | 2002-02-19 | | Release date: | 2002-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined X-ray crystallographic structure of the poliovirus 3C gene product.
J.Mol.Biol., 273, 1997
|
|
1M04
 
 | | Mutant Streptomyces plicatus beta-hexosaminidase (D313N) in complex with product (GlcNAc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Williams, S.J, Mark, B.L, Vocadlo, D.J, James, M.N.G, Withers, S.G. | | Deposit date: | 2002-06-11 | | Release date: | 2002-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical
role in substrate-assisted catalysis by orienting the 2-acetamido group
and stabilizing the transition state.
J.Biol.Chem., 277, 2002
|
|
4APE
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN | | Authors: | Pearl, L.H, Sewell, B.T, Jenkins, J.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1986-06-09 | | Release date: | 1986-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
2QVB
 
 | | Crystal Structure of Haloalkane Dehalogenase Rv2579 from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Haloalkane dehalogenase 3 | | Authors: | Mazumdar, P.A, Hulecki, J, Cherney, M.M, Garen, C.R, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-08-08 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | X-ray crystal structure of Mycobacterium tuberculosis haloalkane dehalogenase Rv2579.
Biochim.Biophys.Acta, 1784, 2008
|
|
