8DU0
 
 | |
5V0U
 
 | |
5VT6
 
 | |
5VIR
 
 | |
5W8J
 
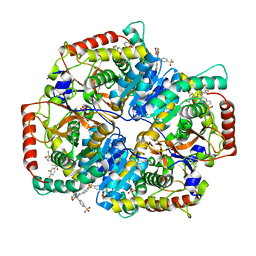 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 29 | | Descriptor: | 2-{3-(3,4-difluorophenyl)-5-hydroxy-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lukacs, C.M, Moulin, A. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
5W8K
 
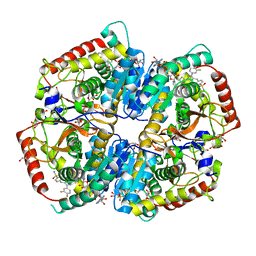 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 29 and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{3-(3,4-difluorophenyl)-5-hydroxy-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, GLYCEROL, ... | | Authors: | Lukacs, C.M, Dranow, D.M. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
5W8I
 
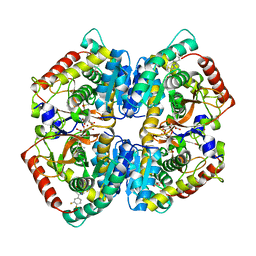 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 23 and Zinc | | Descriptor: | 2-[3-(3,4-difluorophenyl)-5-hydroxy-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Lukacs, C.M, Abendroth, J. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
5W8H
 
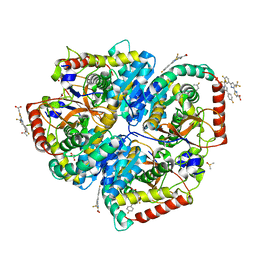 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 11 | | Descriptor: | 2-[3-(4-fluorophenyl)-5-(trifluoromethyl)-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lukacs, C.M, Dranow, D.M. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
4DLP
 
 | |
4G6C
 
 | |
4FUR
 
 | |
4F2N
 
 | |
4F82
 
 | |
4DZ6
 
 | |
4DI6
 
 | |
4H3E
 
 | |
1IOB
 
 | |
6BKW
 
 | | BTK complex with compound 12 | | Descriptor: | GLYCEROL, N-(3-{5-[(1,5-dimethyl-1H-pyrazol-3-yl)amino]-1-methyl-6-oxo-1,6-dihydropyridazin-3-yl}-2,6-difluorophenyl)-4,5,6,7-tetrahydro-1-benzothiophene-2-carboxamide, SULFATE ION, ... | | Authors: | Kiefer, J.R, Eigenbrot, C, Yu, C.L. | | Deposit date: | 2017-11-09 | | Release date: | 2018-11-07 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
6BQA
 
 | | BRD9 bromodomain in complex with 3-(6-(but-3-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-N,N-dimethylbenzamide | | Descriptor: | 3-[6-(but-3-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl]-N,N-dimethylbenzamide, Bromodomain-containing protein 9 | | Authors: | Murray, J.M, Tang, Y. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.031 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
3IUR
 
 | | apPEP_D266Nx+H2H3 opened state | | Descriptor: | GLYCEROL, H2H3 helices from villin headpiece subdomain HP35, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IVM
 
 | | apPEP_WT+PP closed state | | Descriptor: | N-BENZYLOXYCARBONYL-L-PROLYL-L-PROLINAL, prolyl endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-09-01 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IUQ
 
 | | apPEP_D622N+PP closed state | | Descriptor: | GLYCEROL, N-BENZYLOXYCARBONYL-L-PROLYL-L-PROLINAL, Prolyl Endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IUL
 
 | | apPEP_WT1 opened state | | Descriptor: | GLYCEROL, Prolyl endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IEW
 
 | |
3IKE
 
 | |
