6AJN
 
 | | Crystal structure of AtaTR bound with AcCoA | | Descriptor: | ACETYL COENZYME *A, DUF1778 domain-containing protein, N-acetyltransferase | | Authors: | Yashiro, Y, Yamashita, S, Tomita, K. | | Deposit date: | 2018-08-28 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal Structure of the Enterohemorrhagic Escherichia coli AtaT-AtaR Toxin-Antitoxin Complex.
Structure, 27, 2019
|
|
6AJM
 
 | | Crystal structure of apo AtaTR | | Descriptor: | DUF1778 domain-containing protein, N-acetyltransferase, TRIETHYLENE GLYCOL | | Authors: | Yashiro, Y, Yamashita, S, Tomita, K. | | Deposit date: | 2018-08-28 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal Structure of the Enterohemorrhagic Escherichia coli AtaT-AtaR Toxin-Antitoxin Complex.
Structure, 27, 2019
|
|
3IE4
 
 | | b-glucan binding domain of Drosophila GNBP3 defines a novel family of pattern recognition receptor | | Descriptor: | 1,2-ETHANEDIOL, Gram-Negative Binding Protein 3, ZINC ION | | Authors: | Mishima, Y, Coste, F, Kellenberger, C, Roussel, A. | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The N-terminal domain of drosophila gram-negative binding protein 3 (GNBP3) defines a novel family of fungal pattern recognition receptors
To be Published
|
|
7F8D
 
 | | Malate Dehydrogenase from Geobacillus stearothermophilus (gs-MDH) G218Y mutant | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shimozawa, Y, Himiyama, T, Nakamura, T, Nishiya, Y. | | Deposit date: | 2021-07-02 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Increasing loop flexibility affords low-temperature adaptation of a moderate thermophilic malate dehydrogenase from Geobacillus stearothermophilus.
Protein Eng.Des.Sel., 34, 2021
|
|
1PFK
 
 | |
6KV0
 
 | | Ferredoxin I from C. reinhardtii, high X-ray dose | | Descriptor: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-09-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
6LK1
 
 | | Ultrahigh resolution X-ray structure of Ferredoxin I from C. reinhardtii | | Descriptor: | BENZAMIDINE, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-12-17 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
6KUM
 
 | | Ferredoxin I from C. reinhardtii, low X-ray dose | | Descriptor: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-09-02 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
8JX3
 
 | | alpha-Hemolysin(G122S/K147R/K237C)-SpyTag/SpyCatcher head to head 14-mer | | Descriptor: | alpha hemolysin fused with spy-catcher, alpha hemolysin fused with spy-tag | | Authors: | Ishii, Y, Naito, K, Yokoyama, T, Tanaka, Y, Matsuura, T. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | alpha-Hemolysin(G122S/K147R)-SpyTag/SpyCatcher head to head 14-mer
To Be Published
|
|
8JX2
 
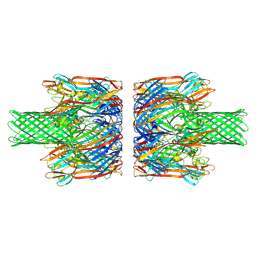 | | alpha-Hemolysin(G122S/K147R)-SpyTag/SpyCatcher head to head 14-mer | | Descriptor: | alpha hemolysin fused with spy-catcher, alpha hemolysin fused with spy-tag | | Authors: | Ishii, Y, Naito, K, Yokoyama, T, Tanaka, Y, Matsuura, T. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | alpha-Hemolysin(G122S/K147R)-SpyTag/SpyCatcher head to head 14-mer
To Be Published
|
|
6KS2
 
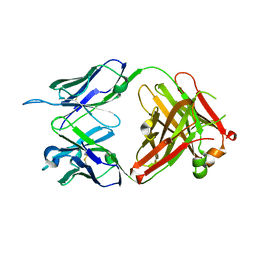 | | Structure of anti-Ghrelin receptor antibody | | Descriptor: | Fab7881 Heavy Chain (FabH), Fab7881 Light Chain (FabL) | | Authors: | Shiimura, Y, Horita, S, Asada, H, Hirata, K, Iwata, S, Kojima, M. | | Deposit date: | 2019-08-23 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structure of an antagonist-bound ghrelin receptor reveals possible ghrelin recognition mode.
Nat Commun, 11, 2020
|
|
6KO5
 
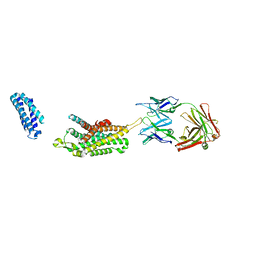 | | Complex structure of Ghrelin receptor with Fab | | Descriptor: | 6-(4-bromanyl-2-fluoranyl-phenoxy)-2-methyl-3-[[(3~{S})-1-propan-2-ylpiperidin-3-yl]methyl]pyrido[3,2-d]pyrimidin-4-one, Chimera of Soluble cytochrome b562 and Growth hormone secretagogue receptor type 1, Fab7881 Heavy Chain, ... | | Authors: | Shiimura, Y, Horita, S, Asada, H, Hirata, K, Iwata, S, Kojima, M. | | Deposit date: | 2019-08-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of an antagonist-bound ghrelin receptor reveals possible ghrelin recognition mode.
Nat Commun, 11, 2020
|
|
4GPG
 
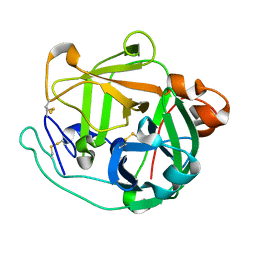 | | X/N joint refinement of Achromobacter Lyticus Protease I free form at pD8.0 | | Descriptor: | Protease 1 | | Authors: | Ohnishi, Y, Yamada, T, Kurihara, K, Tanaka, I, Sakiyama, F, Masaki, T, Niimura, N. | | Deposit date: | 2012-08-21 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-30 | | Method: | NEUTRON DIFFRACTION (1.895 Å), X-RAY DIFFRACTION | | Cite: | Neutron and X-ray crystallographic analysis of Achromobacter protease I at pD 8.0: protonation states and hydration structure in the free-form.
Biochim.Biophys.Acta, 1834, 2013
|
|
4GA4
 
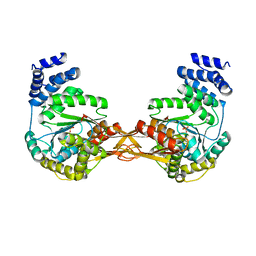 | | Crystal structure of AMP phosphorylase N-terminal deletion mutant | | Descriptor: | PHOSPHATE ION, Putative thymidine phosphorylase | | Authors: | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
4GA5
 
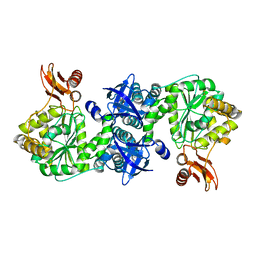 | | Crystal structure of AMP phosphorylase C-terminal deletion mutant in the apo-form | | Descriptor: | Putative thymidine phosphorylase | | Authors: | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2012-07-25 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
8ZYO
 
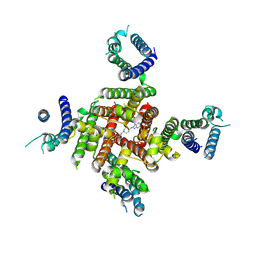 | | Cryo-EM Structure of astemizole-bound hERG Channel | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
8ZYQ
 
 | | Cryo-EM Structure of pimozide-bound hERG Channel | | Descriptor: | 3-[1-[4,4-bis(4-fluorophenyl)butyl]piperidin-4-yl]-1~{H}-benzimidazol-2-one, Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
8ZYP
 
 | | Cryo-EM Structure of E-4031-bound hERG Channel | | Descriptor: | Potassium voltage-gated channel subfamily H member 2, ~{N}-[4-[1-[2-(6-methylpyridin-2-yl)ethyl]piperidin-4-yl]carbonylphenyl]methanesulfonamide | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
8ZYN
 
 | | Cryo-EM Structure of inhibitor-free hERG Channel | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
7WLR
 
 | | Cryo-EM structure of the nucleosome containing Komagataella pastoris histones | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B, ... | | Authors: | Fukushima, Y, Hatazawa, S, Hirai, S, Kujirai, T, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-01-13 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural and biochemical analyses of the nucleosome containing Komagataella pastoris histones.
J.Biochem., 172, 2022
|
|
8J6V
 
 | |
6LB2
 
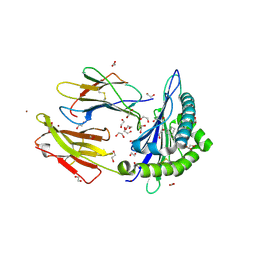 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with mono-acyl glycerol | | Descriptor: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shima, Y, Morita, D. | | Deposit date: | 2019-11-13 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69380951 Å) | | Cite: | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
6LAH
 
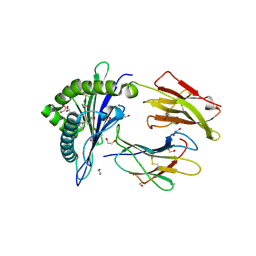 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with lysophosphatidylcholine | | Descriptor: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shima, Y, Morita, D. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
6LAM
 
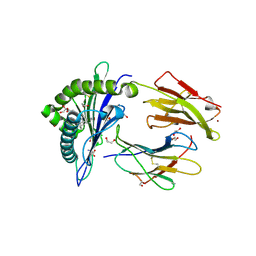 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with lysophosphatidylethanolamine | | Descriptor: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Shima, Y, Morita, D. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
6LN3
 
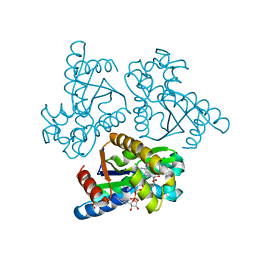 | | Crystal structure of adenylate kinase from an extremophilic archaeon Aeropyrum pernix with ATP and AMP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shibanuma, Y, Nemoto, N, Yamamoto, N, Sampei, G, Kawai, G. | | Deposit date: | 2019-12-28 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of adenylate kinase from an extremophilic archaeon Aeropyrum pernix with ATP and AMP.
J.Biochem., 168, 2020
|
|
