5U4W
 
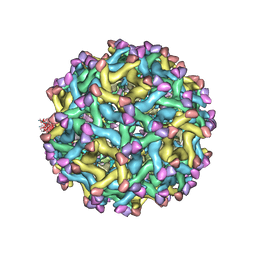 | | Cryo-EM Structure of Immature Zika Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, E protein, M protein, ... | | Authors: | Mangala Prasad, V, Miller, A.S, Klose, T, Sirohi, D, Buda, G, Jiang, W, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2016-12-06 | | Release date: | 2017-01-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Structure of the immature Zika virus at 9 angstrom resolution.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5UHY
 
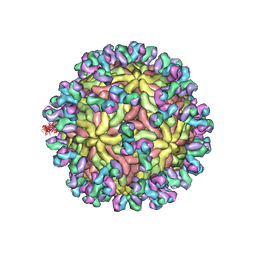 | | A Human Antibody Against Zika Virus Crosslinks the E Protein to Prevent Infection | | Descriptor: | ZV67 Fab chain 1, ZV67 Fab chain 2, envelope protein | | Authors: | Hasan, S.S, Miller, A, Sapparapu, G, Fernandez, E, Klose, T, Long, F, Fokine, A, Porta, J.C, Jiang, W, Diamond, M.S, Crowe Jr, J.E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2017-01-12 | | Release date: | 2017-03-29 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | A human antibody against Zika virus crosslinks the E protein to prevent infection.
Nat Commun, 8, 2017
|
|
1J8S
 
 | | PAPG ADHESIN RECEPTOR BINDING DOMAIN-UNBOUND FORM | | Descriptor: | PYELONEPHRITIC ADHESIN | | Authors: | Dodson, K.W, Pinkner, J.S, Rose, T, Magnusson, G, Hultgren, S.J, Waksman, G. | | Deposit date: | 2001-05-22 | | Release date: | 2001-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor.
Cell(Cambridge,Mass.), 105, 2001
|
|
1IS0
 
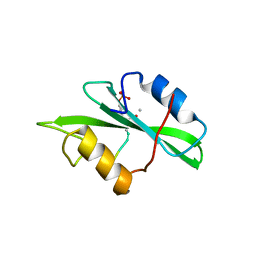 | | Crystal Structure of a Complex of the Src SH2 Domain with Conformationally Constrained Peptide Inhibitor | | Descriptor: | AY0 GLU GLU ILE peptide, Tyrosine-protein kinase transforming protein SRC | | Authors: | Davidson, J.P, Lubman, O, Rose, T, Waksman, G, Martin, S.F. | | Deposit date: | 2001-11-02 | | Release date: | 2002-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calorimetric and structural studies of 1,2,3-trisubstituted cyclopropanes as conformationally constrained peptide inhibitors of Src SH2 domain binding.
J.Am.Chem.Soc., 124, 2002
|
|
1J8R
 
 | | BINARY COMPLEX OF THE PAPG RECEPTOR-BINDING DOMAIN BOUND TO GBO4 RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, PYELONEPHRITIC ADHESIN | | Authors: | Dodson, K.W, Pinkner, J.S, Rose, T, Magnusson, G, Hultgren, S.J, Waksman, G, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-22 | | Release date: | 2001-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor.
Cell(Cambridge,Mass.), 105, 2001
|
|
2D44
 
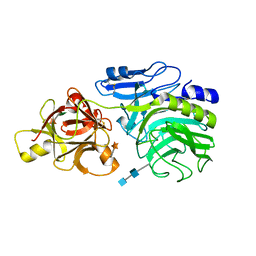 | | Crystal structure of arabinofuranosidase complexed with arabinofuranosyl-alpha-1,2-xylobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranose-(1-2)-alpha-D-xylopyranose-(1-4)-alpha-D-xylopyranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Miwa, Y, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2005-10-07 | | Release date: | 2006-09-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The family 42 carbohydrate-binding module of family 54 alpha-L-arabinofuranosidase specifically binds the arabinofuranose side chain of hemicellulose
Biochem.J., 399, 2006
|
|
2D43
 
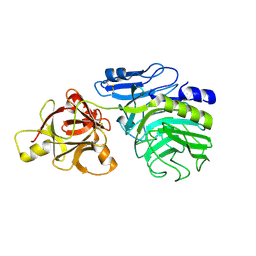 | | Crystal structure of arabinofuranosidase complexed with arabinotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, alpha-L-arabinofuranosidase B | | Authors: | Miyanaga, A, Koseki, T, Miwa, Y, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2005-10-07 | | Release date: | 2006-09-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The family 42 carbohydrate-binding module of family 54 alpha-L-arabinofuranosidase specifically binds the arabinofuranose side chain of hemicellulose
Biochem.J., 399, 2006
|
|
7W71
 
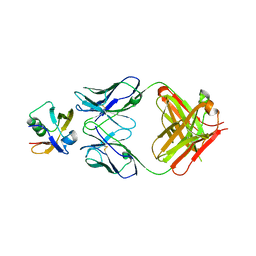 | |
3N4N
 
 | | Insights into the stabilizing contributions of a bicyclic cytosine analogue: crystal structures of DNA duplexes containing 7,8-dihydropyrido[2,3-d]pyrimidin-2-one | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*A)-3', 5'-D(P*TP*TP*(B7C)P*GP*CP*G)-3' | | Authors: | Takenaka, A, Juan, E.C.M, Shimizu, S. | | Deposit date: | 2010-05-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Insights into the DNA stabilizing contributions of a bicyclic cytosine analogue: crystal structures of DNA duplexes containing 7,8-dihydropyrido [2,3-d]pyrimidin-2-one.
Nucleic Acids Res., 2010
|
|
5VU2
 
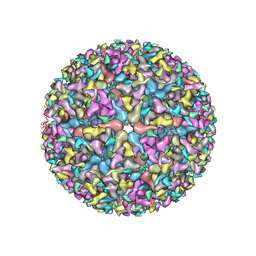 | |
6NCL
 
 | |
9CVG
 
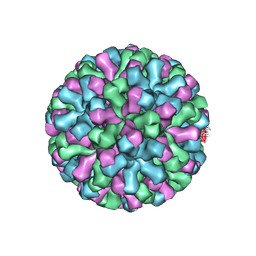 | |
9CVF
 
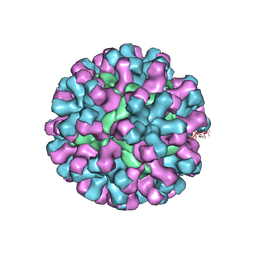 | |
9CVE
 
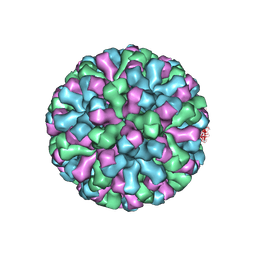 | |
8H1L
 
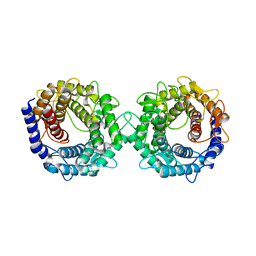 | | Crystal structure of glucose-2-epimerase in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | Descriptor: | N-acylglucosamine 2-epimerase, sorbitol | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1K
 
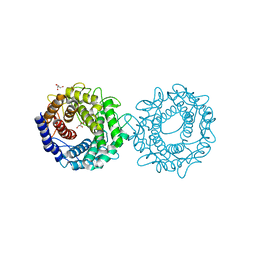 | | Crystal structure of glucose-2-epimerase from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, GLYCEROL, N-acylglucosamine 2-epimerase | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1M
 
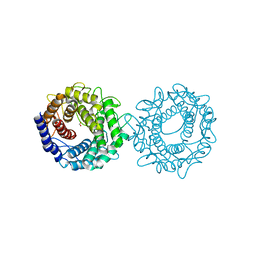 | | Crystal structure of glucose-2-epimerase mutant_D254A from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, N-acylglucosamine 2-epimerase | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H1N
 
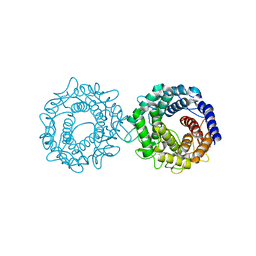 | | Crystal structure of glucose-2-epimerase mutant_D254A in complex with D-Glucitol from Runella slithyformis Runsl_4512 | | Descriptor: | FORMIC ACID, N-acylglucosamine 2-epimerase, sorbitol | | Authors: | Wang, H, Sun, X.M, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural insights into the substrate specificity and activity of a novel mannose 2-epimerase from Runella slithyformis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5H7L
 
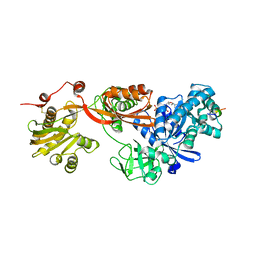 | | Complex of Elongation factor 2-50S ribosomal protein L12 | | Descriptor: | 50S ribosomal protein L12, Elongation factor 2, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Tanzawa, T, Kato, K, Uchiumi, T, Yao, M. | | Deposit date: | 2016-11-18 | | Release date: | 2018-02-21 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The C-terminal helix of ribosomal P stalk recognizes a hydrophobic groove of elongation factor 2 in a novel fashion
Nucleic Acids Res., 46, 2018
|
|
5H7J
 
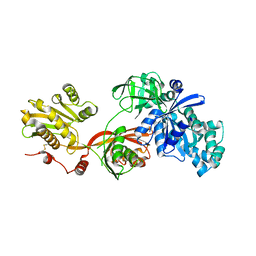 | | Crystal structure of Elongation factor 2 | | Descriptor: | Elongation factor 2, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Tanzawa, T, Kato, K, Uchiumi, T, Yao, M. | | Deposit date: | 2016-11-18 | | Release date: | 2018-02-21 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The C-terminal helix of ribosomal P stalk recognizes a hydrophobic groove of elongation factor 2 in a novel fashion
Nucleic Acids Res., 46, 2018
|
|
5H7K
 
 | | Crystal structure of Elongation factor 2 GDP-form | | Descriptor: | Elongation factor 2, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Tanzawa, T, Kato, K, Uchiumi, T, Yao, M. | | Deposit date: | 2016-11-18 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | The C-terminal helix of ribosomal P stalk recognizes a hydrophobic groove of elongation factor 2 in a novel fashion
Nucleic Acids Res., 46, 2018
|
|
3AX3
 
 | |
3AX2
 
 | | Crystal structure of rat TOM20-ALDH presequence complex: a disulfide-tethered complex with a non-optimized, long linker | | Descriptor: | Aldehyde dehydrogenase, mitochondrial, Mitochondrial import receptor subunit TOM20 homolog, ... | | Authors: | Saitoh, T, Maita, Y, Kohda, D. | | Deposit date: | 2011-03-28 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic snapshots of tom20-mitochondrial presequence interactions with disulfide-stabilized peptides.
Biochemistry, 50, 2011
|
|
3AX5
 
 | | Crystal structure of rat TOM20-ALDH presequence complex: A complex (form1) between Tom20 and a disulfide-bridged presequence peptide containing D-Cys and L-Cys at the i and i+3 positions. | | Descriptor: | Aldehyde dehydrogenase, mitochondrial, Mitochondrial import receptor subunit TOM20 homolog, ... | | Authors: | Saitoh, T, Maita, Y, Kohda, D. | | Deposit date: | 2011-03-29 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic snapshots of tom20-mitochondrial presequence interactions with disulfide-stabilized peptides.
Biochemistry, 50, 2011
|
|
5WLG
 
 | | Crystal Structure of H-2Db with the GAP501 peptide (SQL) | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GAP50 peptide, ... | | Authors: | Gras, S, Farenc, C, Josephs, T, Rossjohn, J. | | Deposit date: | 2017-07-26 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A T Cell Receptor Locus Harbors a Malaria-Specific Immune Response Gene.
Immunity, 47, 2017
|
|
