5MA9
 
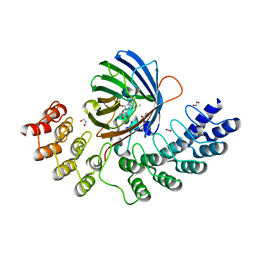 | | GFP-binding DARPin fusion gc_R11 | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, R11 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MC9
 
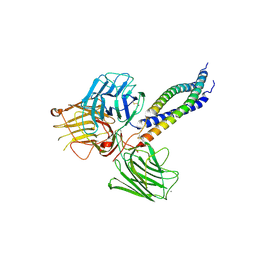 | |
6R9Z
 
 | |
1O6E
 
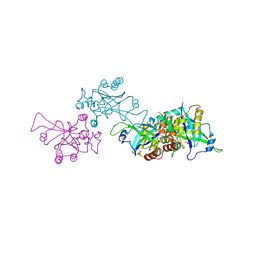 | | Epstein-Barr virus protease | | Descriptor: | CAPSID PROTEIN P40, PHOSPHORYLISOPROPANE | | Authors: | Buisson, M, Hernandez, J, Lascoux, D, Schoehn, G, Forest, E, Arlaud, G, Seigneurin, J, Ruigrok, R.W.H, Burmeister, W.P. | | Deposit date: | 2002-09-13 | | Release date: | 2002-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the Epstein-Barr Virus Protease Shows Rearrangement of the Processed C Terminus
J.Mol.Biol., 324, 2002
|
|
2I0T
 
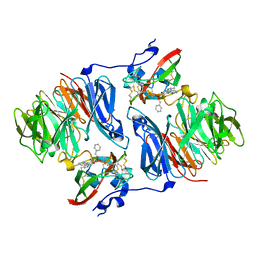 | |
6RAI
 
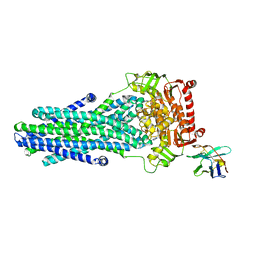 | | Heterodimeric ABC exporter TmrAB in ATP-bound outward-facing occluded conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding and permease protein, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
1O94
 
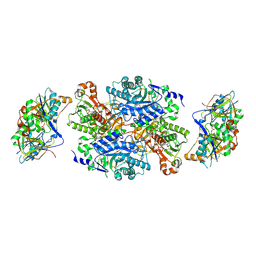 | | Ternary complex between trimethylamine dehydrogenase and electron transferring flavoprotein | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ELECTRON TRANSFER FLAVOPROTEIN ALPHA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
5MEV
 
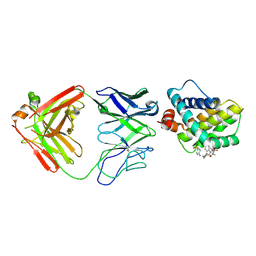 | | MCL1 FAB COMPLEX IN COMPLEX WITH COMPOUND 21 | | Descriptor: | (5~{R},13~{S},17~{S})-5-[(3,4-dichlorophenyl)methyl]-8-methyl-13-[(4-methylsulfonylphenyl)methyl]-1,4,8,12,16-pentazatricyclo[15.8.1.0^{20,25}]hexacosa-20,22,24-triene-3,7,15,26-tetrone, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Hargreaves, D. | | Deposit date: | 2016-11-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors
Acs Med.Chem.Lett., 8, 2017
|
|
5MSG
 
 | |
5M0I
 
 | | Crystal structure of the nuclear complex with She2p and the ASH1 mRNA E3-localization element | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ASH1-E3 element, ... | | Authors: | Edelmann, F.T, Janowski, R, Niessing, D. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Molecular architecture and dynamics of ASH1 mRNA recognition by its mRNA-transport complex.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1OB3
 
 | | Structure of P. falciparum PfPK5 | | Descriptor: | CELL DIVISION CONTROL PROTEIN 2 HOMOLOG | | Authors: | Holton, S, Merckx, A, Burgess, D, Doerig, C, Noble, M, Endicott, J. | | Deposit date: | 2003-01-24 | | Release date: | 2004-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of P. Falciparum Pfpk5 Test the Cdk Regulation Paradigm and Suggest Mechanisms of Small Molecule Inhibition
Structure, 11, 2003
|
|
5M1B
 
 | | Crystal structure of C-terminally tagged apo-UbiD from E. coli | | Descriptor: | 3-octaprenyl-4-hydroxybenzoate carboxy-lyase | | Authors: | White, M, Leys, D. | | Deposit date: | 2016-10-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Oxidative Maturation and Structural Characterization of Prenylated FMN Binding by UbiD, a Decarboxylase Involved in Bacterial Ubiquinone Biosynthesis.
J. Biol. Chem., 292, 2017
|
|
1O6K
 
 | | Structure of activated form of PKB kinase domain S474D with GSK3 peptide and AMP-PNP | | Descriptor: | GLYCOGEN SYNTHASE KINASE-3 BETA, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yang, J, Cron, P, Good, V.M, Thompson, V, Hemmings, B.A, Barford, D. | | Deposit date: | 2002-10-08 | | Release date: | 2002-11-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of an Activated Akt/Protein Kinase B Ternary Complex with Gsk-3 Peptide and AMP-Pnp
Nat.Struct.Biol., 9, 2002
|
|
6RAF
 
 | | Heterodimeric ABC exporter TmrAB in inward-facing narrow conformation under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Anti-vesicular stomatitis virus N VHH, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
1O96
 
 | | Structure of electron transferring flavoprotein for Methylophilus methylotrophus. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ELECTRON TRANSFERRING FLAVOPROTEIN ALPHA-SUBUNIT, ELECTRON TRANSFERRING FLAVOPROTEIN BETA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
2J0E
 
 | | Three dimensional structure and catalytic mechanism of 6- phosphogluconolactonase from Trypanosoma brucei | | Descriptor: | 6-PHOSPHOGLUCONOLACTONASE, MERCURY (II) ION, POTASSIUM ION, ... | | Authors: | Delarue, M, Duclert-Savatier, N, Miclet, E, Haouz, A, Giganti, D, Ouazzani, J, Lopez, P, Nilges, M, Stoven, V. | | Deposit date: | 2006-08-02 | | Release date: | 2007-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three Dimensional Structure and Implications for the Catalytic Mechanism of 6-Phosphogluconolactonase from Trypanosoma Brucei.
J.Mol.Biol., 366, 2007
|
|
5M3V
 
 | | BEAT Fc | | Descriptor: | Ig gamma-1 chain C region, Ig gamma-1 chain C region,Ig gamma-3 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Skegro, D, Stutz, C, Bourquin, F, Blein, S. | | Deposit date: | 2016-10-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Immunoglobulin domain interface exchange as a platform technology for the generation of Fc heterodimers and bispecific antibodies.
J. Biol. Chem., 292, 2017
|
|
1O6L
 
 | | Crystal structure of an activated Akt/protein kinase B (PKB-PIF chimera) ternary complex with AMP-PNP and GSK3 peptide | | Descriptor: | GLYCOGEN SYNTHASE KINASE-3 BETA, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yang, J, Cron, P, Good, V.M, Thompson, V, Hemmings, B.A, Barford, D. | | Deposit date: | 2002-10-08 | | Release date: | 2002-11-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of an Activated Akt/Protein Kinase B Ternary Complex with Gsk-3 Peptide and AMP-Pnp
Nat.Struct.Biol., 9, 2002
|
|
2J1T
 
 | |
6THF
 
 | | Crystal structure of two-domain Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
6T2B
 
 | | Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Glycosyl hydrolase family 109 protein 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Chaberski, E.K, Fredslund, F, Teze, D, Shuoker, B, Kunstmann, S, Karlsson, E.N, Hachem, M.A, Welner, D.H. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Catalytic Acid-Base in GH109 Resides in a Conserved GGHGG Loop and Allows for Comparable alpha-Retaining and beta-Inverting Activity in an N-Acetylgalactosaminidase from Akkermansia muciniphila
Acs Catalysis, 2020
|
|
1OF3
 
 | | Structural and thermodynamic dissection of specific mannan recognition by a carbohydrate-binding module, TmCBM27 | | Descriptor: | BETA-MANNOSIDASE, CALCIUM ION | | Authors: | Boraston, A.B, Revett, T.J, Boraston, C.M, Nurizzo, D, Davies, G.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Dissection of Specific Mannan Recognition by a Carbohydrate Binding Module, Tmcbm27
Structure, 11, 2003
|
|
1OAU
 
 | | Fv Structure of the IgE SPE-7 in complex with DNP-Ser (immunising hapten) | | Descriptor: | 2,4-DINITROPHENOL, IMIDAZOLE, IMMUNOGLOBULIN E, ... | | Authors: | James, L.C, Roversi, P, Tawfik, D. | | Deposit date: | 2003-01-21 | | Release date: | 2004-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antibody Multispecificity Mediated by Conformational Diversity
Science, 299, 2003
|
|
1OHD
 
 | | structure of cdc14 in complex with tungstate | | Descriptor: | CDC14B2 PHOSPHATASE, TUNGSTATE(VI)ION | | Authors: | Gray, C.H, Good, V.M, Tonks, N.K, Barford, D. | | Deposit date: | 2003-05-24 | | Release date: | 2003-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of the Cell Cycle Protein Cdc14 Reveals a Proline-Directed Protein Phosphatase
Embo J., 22, 2003
|
|
5LG0
 
 | | Solution NMR structure of Tryptophan to Alanine mutant of Arkadia RING domain. | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Birkou, M, Chasapis, C.T, Loutsidou, A.K, Bentrop, D, Lelli, M, Herrmann, T, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2016-07-05 | | Release date: | 2017-06-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Residue Specific Insight into the Arkadia E3 Ubiquitin Ligase Activity and Conformational Plasticity.
J. Mol. Biol., 429, 2017
|
|
