6RCP
 
 | |
6RD3
 
 | |
6RCK
 
 | | Crystal structure of the OmpK36 GD insertion chimera from Klebsiella pneumonia | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Beis, K, Romano, M, Kwong, J. | | Deposit date: | 2019-04-11 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | OmpK36-mediated Carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo.
Nat Commun, 10, 2019
|
|
1A3A
 
 | | CRYSTAL STRUCTURE OF IIA MANNITOL FROM ESCHERICHIA COLI | | Descriptor: | MANNITOL-SPECIFIC EII | | Authors: | Van Montfort, R.L.M, Pijning, T, Kalk, K.H, Hangyi, I, Kouwijzer, M.L.C.E, Robillard, G.T, Dijkstra, B.W. | | Deposit date: | 1998-01-19 | | Release date: | 1998-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the Escherichia coli phosphotransferase IIAmannitol reveals a novel fold with two conformations of the active site.
Structure, 6, 1998
|
|
3ZPY
 
 | | Crystal structure of the marine PL7 alginate lyase AlyA1 from Zobellia galactanivorans | | Descriptor: | ALGINATE LYASE, FAMILY PL7, SODIUM ION | | Authors: | Thomas, F, Jeudy, A, Michel, G, Czjzek, M. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Comparative Characterization of Two Marine Alginate Lyases from Zobellia Galactanivorans Reveals Distinct Modes of Action and Exquisite Adaptation to Their Natural Substrate.
J.Biol.Chem., 288, 2013
|
|
7UMJ
 
 | | Crystal structure of recombinant Solieria filiformis lectin (rSfL) | | Descriptor: | Lectin SfL-1, SULFATE ION | | Authors: | Chaves, R.P, Bezerra, E.H.S, da Silva, F.M.S, Carneiro, R.F, Sampaio, A.H, Rocha, B.A.M, Nagano, C.S. | | Deposit date: | 2022-04-07 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural study and antimicrobial and wound healing effects of lectin from Solieria filiformis (Kutzing) P.W.Gabrielson.
Biochimie, 214, 2023
|
|
4BE3
 
 | | crystal structure of the exolytic PL7 alginate lyase AlyA5 from Zobellia galactanivorans | | Descriptor: | ALGINATE LYASE, FAMILY PL7, D(-)-TARTARIC ACID, ... | | Authors: | Thomas, F, Jeudy, A, Michel, G, Czjzek, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Comparative Characterization of Two Marine Alginate Lyases from Zobellia Galactanivorans Reveals Distinct Modes of Action and Exquisite Adaptation to Their Natural Substrate.
J.Biol.Chem., 288, 2013
|
|
6MKQ
 
 | | Carbapenemase VCC-1 bound to avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Mark, B.L, Vadlamani, G. | | Deposit date: | 2018-09-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for the Potent Inhibition of the Emerging Carbapenemase VCC-1 by Avibactam.
Antimicrob. Agents Chemother., 63, 2019
|
|
6SXC
 
 | | Crystal structure of the Voltage-Gated Sodium Channel NavMs (F208L) in complex with 4-hydroxytamoxifen (2.5 Angstrom resolution) | | Descriptor: | 4-HYDROXYTAMOXIFEN, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2019-09-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
6SXF
 
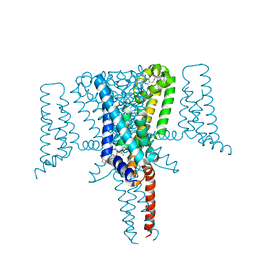 | | Crystal Structure of the Voltage-Gated Sodium Channel NavMs (F208L) in complex with Tamoxifen (2.8 Angstrom resolution) | | Descriptor: | (Z)-2-[4-(1,2)-DIPHENYL-1-BUTENYL)-PHENOXY]-N,N-DIMETHYLETHANAMINE, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2019-09-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.839 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
6SX7
 
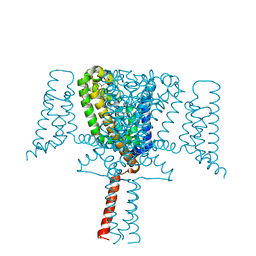 | | Crystal Structure of the Voltage-Gated Sodium Channel NavMs (F208L) (2.2 Angstrom resolution) | | Descriptor: | 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2019-09-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
6SXG
 
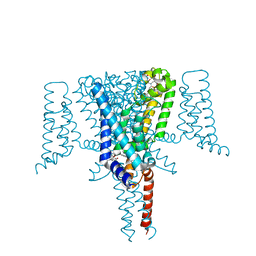 | | Crystal Structure of the Voltage-Gated Sodium Channel NavMs in complex with 4-hydroxytamoxifen (2.4 Angstrom resolution) | | Descriptor: | 4-HYDROXYTAMOXIFEN, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2019-09-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
6HXA
 
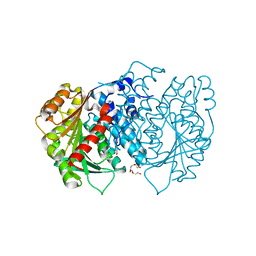 | | AntI from P. luminescens catalyses terminal polyketide shortening in the biosynthesis of anthraquinones | | Descriptor: | AntI, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Zhou, Q, Braeuer, A, Grammbitter, G, Schmalhofer, M, Saura, P, Adihou, H, Kaila, V.R.I, Groll, M, Bode, H. | | Deposit date: | 2018-10-16 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular mechanism of polyketide shortening in anthraquinone biosynthesis ofPhotorhabdus luminescens.
Chem Sci, 10, 2019
|
|
6SXE
 
 | | Crystal Structure of the Voltage-Gated Sodium Channel NavMs (F208L) in complex with Endoxifen (2.6 Angstrom resolution) | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, Endoxifen, HEGA-10, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2019-09-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
6SX5
 
 | | Full length Mutant Open-form Sodium Channel NavMs | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2019-09-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
5FP5
 
 | | Structure of cyclin-dependent kinase 2 with small-molecule ligand 4- fluorobenzoic acid (AT222) in an alternate binding site. | | Descriptor: | 4-fluorobenzoic acid, ACETYL GROUP, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Jhoti, H, Ludlow, R.F, O'Reilly, M, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FP6
 
 | | Structure of cyclin-dependent kinase 2 with small-molecule ligand 3-(4,7-dichloro-1H-indol-3-yl)prop-2-yn-1-ol (AT17833) in an alternate binding site. | | Descriptor: | 3-(4,7-dichloro-1H-indol-3-yl)prop-2-yn-1-ol, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Jhoti, H, Ludlow, R.F, O'Reilly, M, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7PZF
 
 | |
7Q3T
 
 | |
5JUH
 
 | | Crystal structure of C-terminal domain (RV) of MpAFP | | Descriptor: | Antifreeze protein, CALCIUM ION | | Authors: | Guo, S. | | Deposit date: | 2016-05-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5K8G
 
 | |
5J6Y
 
 | | Crystal structure of PA14 domain of MpAFP Antifreeze protein | | Descriptor: | Antifreeze protein, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Guo, S. | | Deposit date: | 2016-04-05 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5IRB
 
 | | Structural insight into host cell surface retention of a 1.5-MDa bacterial ice-binding adhesin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Guo, S, Phippen, S, Campbell, R, Davies, P. | | Deposit date: | 2016-03-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5MRI
 
 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with Triazolone 18 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin, ... | | Authors: | Andersen, J.L, Strandbygaard, D, Thirup, S. | | Deposit date: | 2016-12-23 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The identification of novel acid isostere based inhibitors of the VPS10P family sorting receptor Sortilin.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5MRH
 
 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with Triazolone 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(3-methylbutyl)-4~{H}-1,2,3-triazol-5-one, Sortilin, ... | | Authors: | Andersen, J.L, Strandbygaard, D, Thirup, S. | | Deposit date: | 2016-12-23 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The identification of novel acid isostere based inhibitors of the VPS10P family sorting receptor Sortilin.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
