3CVM
 
 | |
1ZNJ
 
 | | INSULIN, MONOCLINIC CRYSTAL FORM | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Turkenburg, M.G.W, Whittingham, J.L, Turkenburg, J.P, Dodson, G.G, Derewenda, U, Smith, G.D, Dodson, E.J, Derewenda, Z.S, Xiao, B. | | Deposit date: | 1997-09-23 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure Determination and Refinement of Two Crystal Forms of Native Insulins
To be Published
|
|
6RCV
 
 | | PfRH5 bound to monoclonal antibodies R5.011 and R5.016 | | Descriptor: | R5.011 heavy chain, R5.011 light chain, R5.016 heavy chain, ... | | Authors: | Alanine, D.W.G, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (3.582 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
6RCQ
 
 | | PfRH5-binding monoclonal antibody R5.011 | | Descriptor: | ACETATE ION, CACODYLATE ION, R5.011 heavy chain, ... | | Authors: | Alanine, D.W.G, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
6RCS
 
 | | PfRH5-binding monoclonal antibody R5.016 | | Descriptor: | ACETATE ION, R5.016 heavy chain, R5.016 light chain, ... | | Authors: | Alanine, D.W.G, Jamwal, A, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
7PDU
 
 | |
7PCZ
 
 | | Functional and structural characterization of redox sensitive superfolder green fluorescent protein and variants | | Descriptor: | ETHANOL, GLYCEROL, Green fluorescent protein | | Authors: | Fritz-Wolf, K, Heimsch, K.C, Schuh, A.K, Becker, K. | | Deposit date: | 2021-08-04 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Function of Redox-Sensitive Superfolder Green Fluorescent Protein Variant.
Antioxid.Redox Signal., 37, 2022
|
|
7PCA
 
 | | Functional and structural characterization of redox sensitive superfolder green fluorescent protein and variants | | Descriptor: | ETHANOL, FORMAMIDE, GLYCEROL, ... | | Authors: | Fritz-Wolf, K, Heimsch, K.C, Schuh, A.K, Becker, K. | | Deposit date: | 2021-08-03 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure and Function of Redox-Sensitive Superfolder Green Fluorescent Protein Variant.
Antioxid.Redox Signal., 37, 2022
|
|
7PD0
 
 | | Functional and structural characterization of redox sensitive superfolder green fluorescent protein and variants | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Green fluorescent protein, ... | | Authors: | Fritz-Wolf, K, Heimsch, K.C, Schuh, A.K, Becker, K. | | Deposit date: | 2021-08-04 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of Redox-Sensitive Superfolder Green Fluorescent Protein Variant.
Antioxid.Redox Signal., 37, 2022
|
|
6R2S
 
 | |
6RCU
 
 | | PfRH5 bound to monoclonal antibodies R5.004 and R5.016 | | Descriptor: | R5.004 heavy chain, R5.004 light chain, R5.016 heavy chain, ... | | Authors: | Alanine, D.W.G, Draper, S.J, Higgins, M.K. | | Deposit date: | 2019-04-11 | | Release date: | 2019-06-26 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Human Antibodies that Slow Erythrocyte Invasion Potentiate Malaria-Neutralizing Antibodies.
Cell, 178, 2019
|
|
1ZNI
 
 | | INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, ZINC ION | | Authors: | Turkenburg, M.G.W, Whittingham, J.L, Dodson, G.G, Dodson, E.J, Xiao, B, Bentley, G.A. | | Deposit date: | 1997-09-23 | | Release date: | 1998-01-28 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Structure of insulin in 4-zinc insulin.
Nature, 261, 1976
|
|
4YXW
 
 | | Bovine heart mitochondrial F1-ATPase inhibited by AMP-PNP and ADP in the presence of thiophosphate. | | Descriptor: | ATP synthase subunit alpha, mitochondrial, ATP synthase subunit beta, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How release of phosphate from mammalian F1-ATPase generates a rotary substep.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5IK2
 
 | | Caldalaklibacillus thermarum F1-ATPase (epsilon mutant) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Ferguson, S.A, Cook, G.M, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2016-03-03 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4Z1M
 
 | | Bovine F1-ATPase inhibited by three copies of the inhibitor protein IF1 crystallised in the presence of thiophosphate. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | How release of phosphate from mammalian F1-ATPase generates a rotary substep.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A8E
 
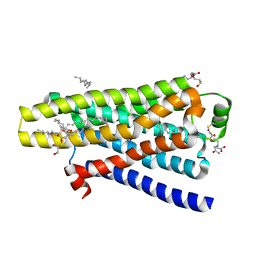 | | thermostabilised beta1-adrenoceptor with rationally designed inverse agonist 7-methylcyanopindolol bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4-[(2S)-3-(tert-butylamino)-2-hydroxypropoxy]-7-methyl-1H-indole-2-carbonitrile, ... | | Authors: | Sato, T, Baker, J.G, Warne, T, Brown, G.A, Congreve, M, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacological Analysis and Structure Determination of 7-Methylcyanopindolol-Bound Beta1-Adrenergic Receptor.
Mol.Pharmacol., 88, 2015
|
|
4TT3
 
 | | The Pathway of Binding of the Intrinsically Disordered Mitochondrial Inhibitor Protein to F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2014-06-19 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TSF
 
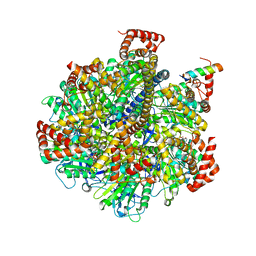 | | The Pathway of Binding of the Intrinsically Disordered Mitochondrial Inhibitor Protein to F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1NBM
 
 | | THE STRUCTURE OF BOVINE F1-ATPASE COVALENTLY INHIBITED WITH 4-CHLORO-7-NITROBENZOFURAZAN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, F1-ATPASE, ... | | Authors: | Orriss, G.L, Leslie, A.G.W, Braig, K, Walker, J.E. | | Deposit date: | 1998-04-30 | | Release date: | 1998-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bovine F1-ATPase covalently inhibited with 4-chloro-7-nitrobenzofurazan: the structure provides further support for a rotary catalytic mechanism.
Structure, 6, 1998
|
|
3FKS
 
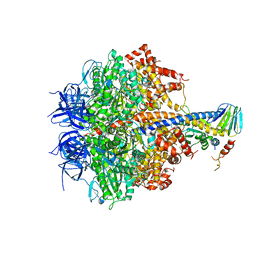 | | Yeast F1 ATPase in the absence of bound nucleotides | | Descriptor: | ATP synthase subunit alpha, mitochondrial, ATP synthase subunit beta, ... | | Authors: | Kabaleeswaran, V, Symersky, J, Shen, H, Walker, J.E, Leslie, A.G.W, Mueller, D.M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.587 Å) | | Cite: | Asymmetric structure of the yeast f1 ATPase in the absence of bound nucleotides.
J.Biol.Chem., 284, 2009
|
|
3ZRY
 
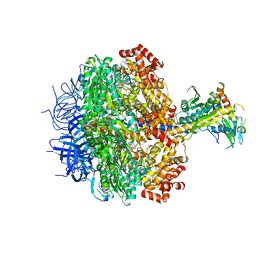 | |
3J6K
 
 | |
4AHC
 
 | |
4AIL
 
 | | Crystal Structure of an Evolved Replicating DNA Polymerase | | Descriptor: | 5'-D(*AP*CP*GP*GP*GP*TP*AP*AP*GP*CP*AP)-3', 5'-D(*TP*GP*CP*TP*TP*AP*CP*DOCP)-3', DNA POLYMERASE | | Authors: | Wynne, S.A, Holliger, P, Leslie, A.G.W. | | Deposit date: | 2012-02-10 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of an Apo and a Binary Complex of an Evolved Archeal B Family DNA Polymerase Capable of Synthesising Highly Cy-Dye Labelled DNA.
Plos One, 8, 2013
|
|
3H5C
 
 | | X-Ray Structure of Protein Z-Protein Z Inhibitor Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein Z-dependent protease inhibitor, Vitamin K-dependent protein Z, ... | | Authors: | Dementiev, A.A, Huang, X, Olson, S.T, Gettins, P.G.W. | | Deposit date: | 2009-04-21 | | Release date: | 2010-04-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Basis for the specificity and activation of the serpin protein Z-dependent proteinase inhibitor (ZPI) as an inhibitor of membrane-associated factor Xa.
J.Biol.Chem., 285, 2010
|
|
