6Z5R
 
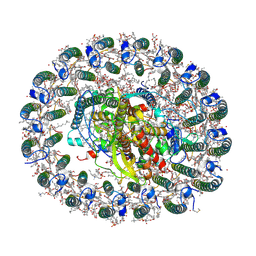 | | RC-LH1(16) complex from Rhodopseudomonas palustris | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, (6~{R},10~{S},14~{R},19~{R},23~{S},24~{E},27~{S},28~{E})-2,6,10,14,19,23,27,31-octamethyldotriaconta-24,28-dien-2-ol, ... | | Authors: | Swainsbury, D.J.K, Qian, P, Hitchcock, A, Hunter, C.N. | | Deposit date: | 2020-05-27 | | Release date: | 2021-01-13 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of Rhodopseudomonas palustris RC-LH1 complexes with open or closed quinone channels.
Sci Adv, 7, 2021
|
|
6Z8E
 
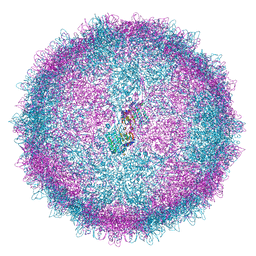 | | Human Picobirnavirus Ht-CP VLP | | Descriptor: | Capsid protein precursor | | Authors: | Ortega-Esteban, A, Mata, C.P, Rodriguez-Espinosa, M.J, Luque, D, Irigoyen, N, Rodriguez, J.M, de Pablo, P.J, Caston, J.R. | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-electron Microscopy Structure, Assembly, and Mechanics Show Morphogenesis and Evolution of Human Picobirnavirus.
J.Virol., 94, 2020
|
|
6Z9G
 
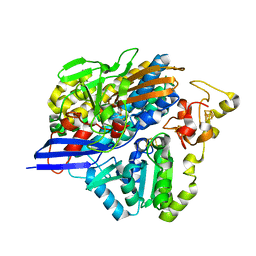 | | Structure of [NiFeSe] hydrogenase G491A variant from Desulfovibrio vulgaris Hildenborough pressurized with Oxygen gas - structure G491A-O2 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE (II) ION, ... | | Authors: | Zacarias, S, Temporao, A, Carpentier, P, van der Linden, P, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Exploring the gas access routes in a [NiFeSe] hydrogenase using crystals pressurized with krypton and oxygen.
J.Biol.Inorg.Chem., 25, 2020
|
|
6Z6U
 
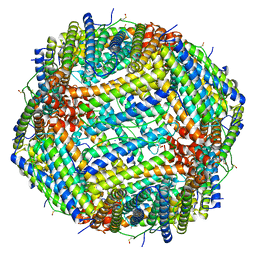 | | 1.25 A structure of human apoferritin obtained from Titan Mono-BCOR microscope | | Descriptor: | Ferritin heavy chain, MAGNESIUM ION, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Paknia, E, Chari, A, Stark, H. | | Deposit date: | 2020-05-29 | | Release date: | 2020-06-24 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (1.25 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
6ZCT
 
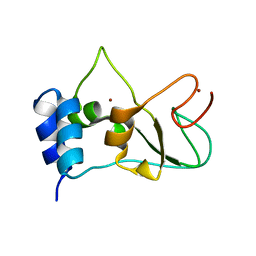 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | ZINC ION, nsp10 | | Authors: | Rogstam, A, Nyblom, M, Christensen, S, Sele, C, Lindvall, T, Rasmussen, A.A, Andre, I, Fisher, S.Z, Knecht, W, Kozielski, F. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
6ZD4
 
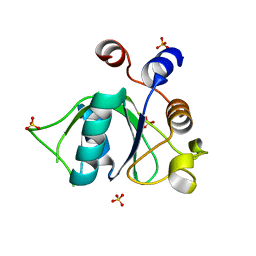 | | Crystal structure of YTHDC1 S378A mutant | | Descriptor: | SULFATE ION, YTH domain containing 1 | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2020-06-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6Z9L
 
 | | Enterococcal PrgA | | Descriptor: | Poly-alanine peptide, PrgA, SULFATE ION | | Authors: | Berntsson, R.P.A, Schmitt, A. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.063 Å) | | Cite: | Enterococcal PrgA Extends Far Outside the Cell and Provides Surface Exclusion to Protect against Unwanted Conjugation.
J.Mol.Biol., 432, 2020
|
|
6ZFJ
 
 | |
6ZGB
 
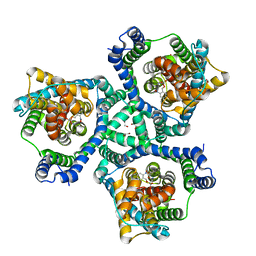 | | glutamate transporter homologue Glttk in complex with a photo cage compound | | Descriptor: | (2~{S},3~{S})-2-azanyl-3-[(2-nitrophenyl)methoxy]butanedioic acid, DI(HYDROXYETHYL)ETHER, Proton/glutamate symporter, ... | | Authors: | Arkhipova, V, Slotboom, D.J, Guskov, A. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Aspects of Photopharmacology: Insight into the Binding of Photoswitchable and Photocaged Inhibitors to the Glutamate Transporter Homologue.
J.Am.Chem.Soc., 143, 2021
|
|
6ZCO
 
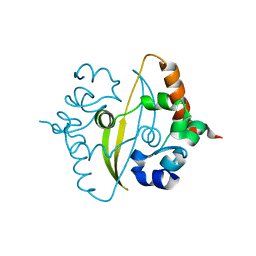 | | Crystal Structure of C-terminal Dimerization Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2, crystal form II | | Descriptor: | Nucleoprotein | | Authors: | Zinzula, L, Basquin, J, Nagy, I, Bracher, A. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.361 Å) | | Cite: | High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus 2.
Biochem.Biophys.Res.Commun., 538, 2021
|
|
6ZCV
 
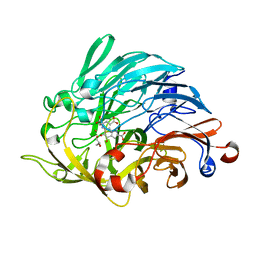 | |
6ZJ9
 
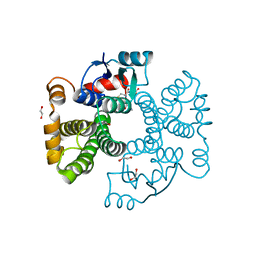 | | Crystal structure of Equus ferus caballus glutathione transferase A3-3 in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase | | Authors: | Skerlova, J, Ismail, A, Lindstrom, H, Sjodin, B, Mannervik, B, Stenmark, P. | | Deposit date: | 2020-06-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of the inhibition of equine glutathione transferase A3-3 by organotin endocrine disrupting pollutants.
Environ Pollut, 268, 2021
|
|
6ZL6
 
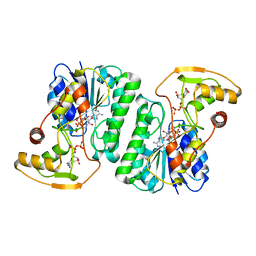 | |
6Z9E
 
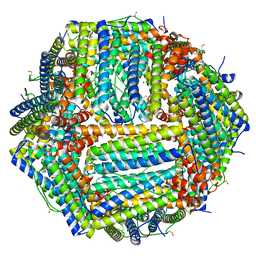 | | 1.55 A structure of human apoferritin obtained from data subset of Titan Mono-BCOR microscope | | Descriptor: | Ferritin heavy chain, SODIUM ION | | Authors: | Yip, K.M, Fischer, N, Paknia, E, Chari, A, Stark, H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (1.55 Å) | | Cite: | Atomic-resolution protein structure determination by cryo-EM.
Nature, 587, 2020
|
|
6ZCM
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_180 | | Descriptor: | 6-[[cyclopropyl-[(7-methoxy-1,3-benzodioxol-5-yl)methyl]amino]methyl]-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6ZD3
 
 | | Crystal structure of YTHDC1 M438A mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, YTH domain containing 1 | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2020-06-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Atomistic and Thermodynamic Analysis of N6-Methyladenosine (m 6 A) Recognition by the Reader Domain of YTHDC1.
J Chem Theory Comput, 17, 2021
|
|
6Z4U
 
 | |
6ZA1
 
 | | Structure of [NiFeSe] hydrogenase G491A variant from Desulfovibrio vulgaris Hildenborough pressurized with Oxygen gas - structure G491A-O2-hd | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, GLYCEROL, ... | | Authors: | Zacarias, S, Temporao, A, Carpentier, P, van der Linden, P, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Exploring the gas access routes in a [NiFeSe] hydrogenase using crystals pressurized with krypton and oxygen.
J.Biol.Inorg.Chem., 25, 2020
|
|
6ZCW
 
 | |
6Z44
 
 | | Crystal structure of the cAMP-dependent protein kinase A in complex with phenol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DIMETHYL SULFOXIDE, PHENOL, ... | | Authors: | Oebbeke, M, Heine, A, Klebe, G. | | Deposit date: | 2020-05-22 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6ZIG
 
 | |
6YWM
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with MES | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Ni, X, Schroeder, M, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6YX5
 
 | | Structure of DrrA from Legionella pneumophilia in complex with human Rab8a | | Descriptor: | MAGNESIUM ION, Multifunctional virulence effector protein DrrA, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Schneider, S, Du, J, von Wrisberg, M.K, Lang, K, Itzen, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Rab1-AMPylation by Legionella DrrA is allosterically activated by Rab1.
Nat Commun, 12, 2021
|
|
6YZA
 
 | |
6ZLN
 
 | | CLK1 bound with GW807982X (Cpd 8) | | Descriptor: | 1,2-ETHANEDIOL, 4-(6-ethoxypyrazolo[1,5-b]pyridazin-3-yl)-~{N}-[3-methoxy-5-(trifluoromethyl)phenyl]pyrimidin-2-amine, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | DFG-1 Residue Controls Inhibitor Binding Mode and Affinity, Providing a Basis for Rational Design of Kinase Inhibitor Selectivity.
J.Med.Chem., 63, 2020
|
|
