7CWU
 
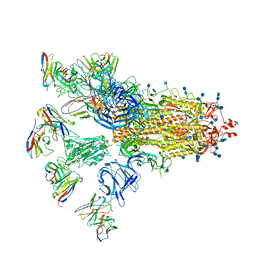 | | SARS-CoV-2 spike proteins trimer in complex with P17 and FC05 Fabs cocktail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-31 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-based development of human antibody cocktails against SARS-CoV-2.
Cell Res., 31, 2021
|
|
5TSU
 
 | | Active conformation for Engineered human cystathionine gamma lyase (E59N, R119L, E339V) to depleting methionine | | Descriptor: | CYSTEINE, Cystathionine gamma-lyase, METHIONINE, ... | | Authors: | Yan, W, Zhang, Y. | | Deposit date: | 2016-10-31 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Snapshots of an Engineered Cystathionine-gamma-lyase Reveal the Critical Role of Electrostatic Interactions in the Active Site.
Biochemistry, 56, 2017
|
|
7CWT
 
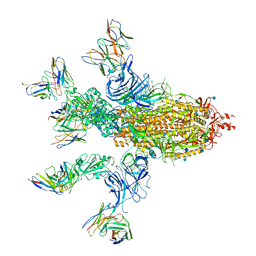 | |
5V12
 
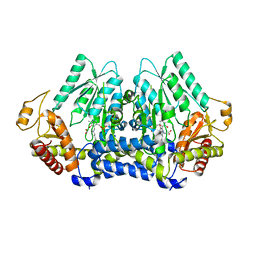 | | Crystal structure of Carbon Sulfoxide lyase, Egt2 Y134F with sulfenic acid intermediate | | Descriptor: | (1S)-1-carboxy-2-[2-(hydroxysulfanyl)-1H-imidazol-4-yl]-N,N,N-trimethylethan-1-aminium, FORMIC ACID, Hercynylcysteine sulfoxide lyase | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2017-03-01 | | Release date: | 2018-03-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Snapshots of C-S Cleavage in Egt2 Reveals Substrate Specificity and Reaction Mechanism.
Cell Chem Biol, 25, 2018
|
|
5GMO
 
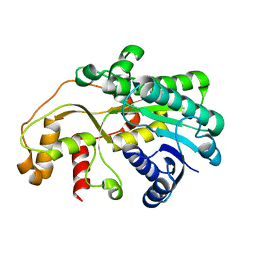 | | X-ray structure of carbonyl reductase SsCR | | Descriptor: | Protein induced by osmotic stress | | Authors: | Shang, Y.P, Yu, H.L, Xu, J.H. | | Deposit date: | 2016-07-14 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Efficient Synthesis of (R)-2-Chloro-1-(2,4-dichlorophenyl)ethanol with a Ketoreductase from Scheffersomyces stipitis CBS 6045
Adv.Synth.Catal., 359, 2017
|
|
6AKS
 
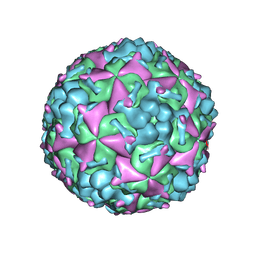 | | Cryo-EM structure of CVA10 mature virus | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AKT
 
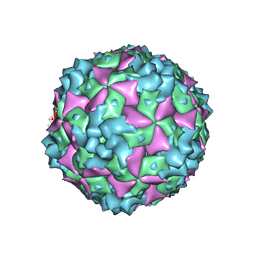 | | Cryo-EM structure of CVA10 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AKU
 
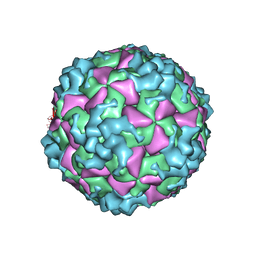 | | Cryo-EM structure of CVA10 empty particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6KW1
 
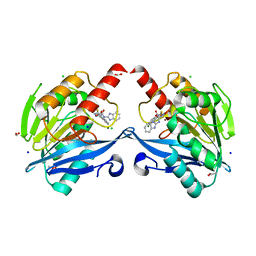 | | The structure of the metallo-beta-lactamase VIM-2 in complex with a triazolylthioacetamide 1b | | Descriptor: | 2-[3-[2-(1H-benzimidazol-2-ylamino)-2-oxidanylidene-ethyl]sulfanyl-1H-1,2,4-triazol-5-yl]benzoic acid, ACETATE ION, Beta-lactamase class B VIM-2, ... | | Authors: | Yang, K.W, Xiang, Y. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.775212 Å) | | Cite: | Kinetic, Thermodynamic, and Crystallographic Studies of 2-Triazolylthioacetamides as Verona Integron-Encoded Metallo-beta-Lactamase 2 (VIM-2) Inhibitor.
Biomolecules, 10, 2020
|
|
7VWA
 
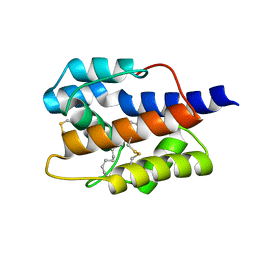 | |
7VW8
 
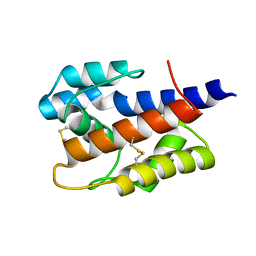 | |
7VW9
 
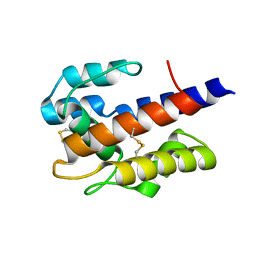 | |
1HZ5
 
 | | CRYSTAL STRUCTURES OF THE B1 DOMAIN OF PROTEIN L FROM PEPTOSTREPTOCOCCUS MAGNUS, WITH A TYROSINE TO TRYPTOPHAN SUBSTITUTION | | Descriptor: | PROTEIN L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-01-23 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the B1 domain of protein L from Peptostreptococcus magnus with a tyrosine to tryptophan substitution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HZ6
 
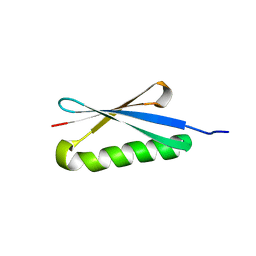 | | CRYSTAL STRUCTURES OF THE B1 DOMAIN OF PROTEIN L FROM PEPTOSTREPTOCOCCUS MAGNUS WITH A TYROSINE TO TRYPTOPHAN SUBSTITUTION | | Descriptor: | PROTEIN L | | Authors: | O'Neill, J.W, Kim, D.E, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-01-23 | | Release date: | 2001-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the B1 domain of protein L from Peptostreptococcus magnus with a tyrosine to tryptophan substitution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1JML
 
 | | Conversion of Monomeric Protein L to an Obligate Dimer by Computational Protein Design | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kuhlman, B, Kim, D.E, Zhang, K.Y.J, Baker, D. | | Deposit date: | 2001-07-19 | | Release date: | 2001-10-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conversion of monomeric protein L to an obligate dimer by computational protein design.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K50
 
 | | A V49A Mutation Induces 3D Domain Swapping in the B1 Domain of Protein L from Peptostreptococcus magnus | | Descriptor: | Protein L | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K53
 
 | | Monomeric Protein L B1 Domain with a G15A Mutation | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K51
 
 | | A G55A Mutation Induces 3D Domain Swapping in the B1 Domain of Protein L from Peptostreptococcus magnus | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K52
 
 | | Monomeric Protein L B1 Domain with a K54G mutation | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
5I1Z
 
 | | Structure of nvPizza2-H16S58 | | Descriptor: | SULFATE ION, nvPizza2-H16S58 | | Authors: | Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2016-02-07 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Broken Symmetry: Partial domain swapping in an artificial trimeric protein
To Be Published
|
|
7DG7
 
 | |
7DG9
 
 | | DPBB domain of VCP-like ATPase from Aeropyrum pernix | | Descriptor: | Cell division control protein 48, AAA family, ZINC ION | | Authors: | Yagi, S, Tagami, S. | | Deposit date: | 2020-11-11 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Seven Amino Acid Types Suffice to Create the Core Fold of RNA Polymerase.
J.Am.Chem.Soc., 143, 2021
|
|
7DBO
 
 | |
4FK3
 
 | | B-Raf Kinase V600E Oncogenic Mutant in Complex with PLX3203 | | Descriptor: | N-{2,4-difluoro-3-[(5-pyridin-3-yl-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]phenyl}ethanesulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Zhang, Y, Wang, W, Zhang, K.Y.J. | | Deposit date: | 2012-06-12 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5F53
 
 | |
