6IHI
 
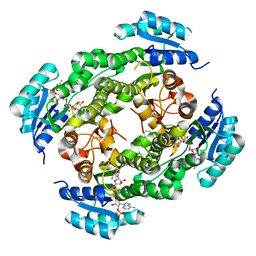 | | Crystal structure of RasADH 3B3/I91V from Ralstonia.sp in complex with NADPH and A6O | | Descriptor: | (2R,3S)-2-ethyl-2-[(2E)-2-(6-methoxy-3,4-dihydro-2H-naphthalen-1-ylidene)ethyl]-3-oxidanyl-cyclopentan-1-one, Alclohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, H.L, Chen, X, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2018-09-30 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Efficient reductive desymmetrization of bulky 1,3-cyclodiketones enabled by structure-guided directed evolution of a carbonyl reductase.
Nat Catal, 2, 2019
|
|
7R75
 
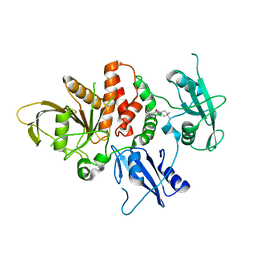 | | Structure of human SHP2 in complex with compound 16 | | Descriptor: | 6-(4-amino-4-methylpiperidin-1-yl)-3-(3-chlorophenyl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Leonard, P.G, Cross, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of 6-[(3 S ,4 S )-4-Amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methyl-3,4-dihydropyrimidin-4-one (IACS-15414), a Potent and Orally Bioavailable SHP2 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7R7D
 
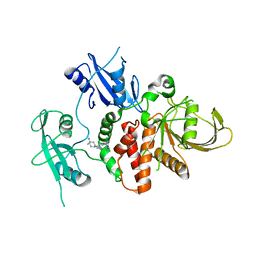 | | Structure of human SHP2 in complex with compound 22 | | Descriptor: | 4-[6-(4-amino-4-methylpiperidin-1-yl)-1H-pyrazolo[3,4-b]pyrazin-3-yl]-3-chloro-N-methylpyridin-2-amine, TETRAETHYLENE GLYCOL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Leonard, P.G, Cross, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 6-[(3 S ,4 S )-4-Amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methyl-3,4-dihydropyrimidin-4-one (IACS-15414), a Potent and Orally Bioavailable SHP2 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7R7L
 
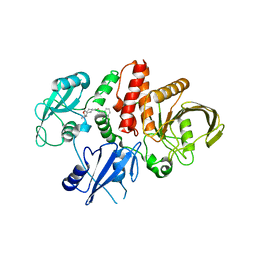 | | Structure of human SHP2 in complex with compound 30 | | Descriptor: | 6-[(3S,4S)-4-amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methylpyrimidin-4(3H)-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Leonard, P.G, Cross, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of 6-[(3 S ,4 S )-4-Amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methyl-3,4-dihydropyrimidin-4-one (IACS-15414), a Potent and Orally Bioavailable SHP2 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7R7I
 
 | | Structure of human SHP2 in complex with compound 27 | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11, [3-(4-amino-4-methylpiperidin-1-yl)-6-(2,3-dichlorophenyl)-5-methylpyrazin-2-yl]methanol | | Authors: | Leonard, P.G, Cross, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of 6-[(3 S ,4 S )-4-Amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methyl-3,4-dihydropyrimidin-4-one (IACS-15414), a Potent and Orally Bioavailable SHP2 Inhibitor.
J.Med.Chem., 64, 2021
|
|
6IHH
 
 | | Crystal structure of RasADH F12 from Ralstonia.sp in complex with NADPH and A6O | | Descriptor: | (2R,3S)-2-ethyl-2-[(2E)-2-(6-methoxy-3,4-dihydro-2H-naphthalen-1-ylidene)ethyl]-3-oxidanyl-cyclopentan-1-one, Alclohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, H.L, Chen, X, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2018-09-30 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efficient reductive desymmetrization of bulky 1,3-cyclodiketones enabled by structure-guided directed evolution of a carbonyl reductase.
Nat Catal, 2, 2019
|
|
3WTF
 
 | | Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Ochi, T, Blundell, T.L. | | Deposit date: | 2014-04-09 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.451 Å) | | Cite: | DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair.
Science, 347, 2015
|
|
3WTD
 
 | | Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Ochi, T, Blundell, T.L. | | Deposit date: | 2014-04-09 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair.
Science, 347, 2015
|
|
5K0M
 
 | | Targeting the PRC2 complex through a novel protein-protein interaction inhibitor of EED | | Descriptor: | (3R,4S)-1-[(1S)-7-fluoro-2,3-dihydro-1H-inden-1-yl]-N,N-dimethyl-4-{4-[4-(methylsulfonyl)piperazin-1-yl]phenyl}pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Bigelow, L.J, Zhu, H, Sun, C. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex.
Nat. Chem. Biol., 13, 2017
|
|
3MZI
 
 | |
7MK2
 
 | | CryoEM Structure of NPR1 | | Descriptor: | Regulatory protein NPR1, ZINC ION | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M, Bartesaghi, A, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
6KZP
 
 | | calcium channel-ligand | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N. | | Deposit date: | 2019-09-25 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of apo and antagonist-bound human Cav3.1.
Nature, 576, 2019
|
|
6KZO
 
 | | membrane protein | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N. | | Deposit date: | 2019-09-25 | | Release date: | 2019-12-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of apo and antagonist-bound human Cav3.1.
Nature, 576, 2019
|
|
4YRV
 
 | | Crystal structure of Anabaena transcription factor HetR complexed with 21-bp DNA from hetP promoter | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*GP*AP*CP*CP*CP*CP*TP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*AP*GP*GP*GP*GP*TP*CP*TP*AP*AP*CP*CP*CP*CP*TP*CP*AP*T)-3'), ... | | Authors: | Hu, H.X, Jiang, Y.L, Zhao, M.X, Zhang, C.C, Chen, Y, Zhou, C.Z. | | Deposit date: | 2015-03-16 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into HetR-PatS interaction involved in cyanobacterial pattern formation
Sci Rep, 5, 2015
|
|
5D2S
 
 | |
4ZMD
 
 | | C domain of staphylococcal protein A mutant - Q9W | | Descriptor: | Immunoglobulin G-binding protein A | | Authors: | Deis, L.N, Oas, T.G. | | Deposit date: | 2015-05-03 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Suppression of conformational heterogeneity at a protein-protein interface.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZXT
 
 | | Complex of ERK2 with catechol | | Descriptor: | AMMONIUM ION, CATECHOL, Mitogen-activated protein kinase 1, ... | | Authors: | Kurinov, I, Malakhova, M. | | Deposit date: | 2015-05-20 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A natural small molecule, catechol, induces c-Myc degradation by directly targeting ERK2 in lung cancer.
Oncotarget, 7, 2016
|
|
5D2Q
 
 | |
5D23
 
 | |
4ZNC
 
 | |
4YNL
 
 | | Crystal structure of the hood domain of Anabaena HetR in complex with the hexapeptide ERGSGR derived from PatS | | Descriptor: | Heterocyst differentiation control protein, Heterocyst inhibition-signaling peptide | | Authors: | Hu, H.X, Jiang, Y.L, Zhao, M.X, Zhang, C.C, Chen, Y, Zhou, C.Z. | | Deposit date: | 2015-03-10 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into HetR-PatS interaction involved in cyanobacterial pattern formation
Sci Rep, 5, 2015
|
|
5DRO
 
 | | Structure of the Aquifex aeolicus LpxC/LPC-011 Complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Najeeb, J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5DRP
 
 | | Structure of the AaLpxC/LPC-023 Complex | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, N~2~-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N-hydroxy-L-isoleucinamide, ... | | Authors: | Najeeb, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5DRR
 
 | | Crystal structure of the Pseudomonas aeruginosa LpxC/LPC-058 complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]benzamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Najeeb, J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
6M53
 
 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, GLYCEROL, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
