3L58
 
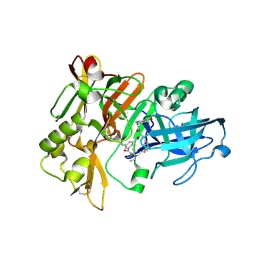 | | Structure of BACE Bound to SCH589432 | | Descriptor: | Beta-secretase 1, N'-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-5-METHYL-N,N-DIPROPYLISOPHTHALAMIDE | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3LPI
 
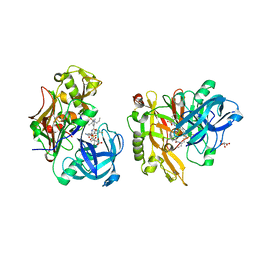 | | Structure of BACE Bound to SCH745132 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylsulfonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L5B
 
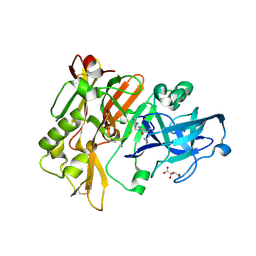 | | Structure of BACE Bound to SCH713601 | | Descriptor: | (2Z,5R)-3-(3-chlorobenzyl)-2-imino-5-methyl-5-(2-methylpropyl)imidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5E
 
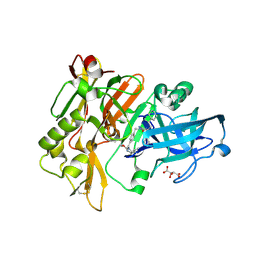 | | Structure of BACE Bound to SCH736062 | | Descriptor: | (4S)-1-(4-{[(2Z,4R)-4-(2-cyclohexylethyl)-4-(cyclohexylmethyl)-2-imino-5-oxoimidazolidin-1-yl]methyl}benzyl)-4-propylimidazolidin-2-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3LPK
 
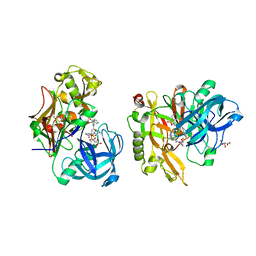 | | Structure of BACE Bound to SCH747123 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-{(2R)-4-[(3-methylphenyl)sulfonyl]piperazin-2-yl}ethyl]-3-{[(2R)-2-(methoxymethyl)pyrrolidin-1-yl]carbonyl}-5-methylbenzamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KKO
 
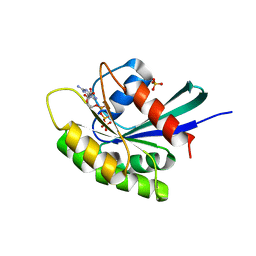 | | Crystal structure of M-Ras P40D/D41E/L51R in complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein M-Ras, ... | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3KKM
 
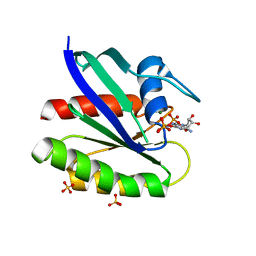 | | Crystal structure of H-Ras T35S in complex with GppNHp | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3KKP
 
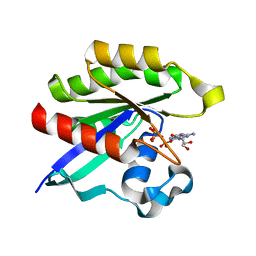 | | Crystal structure of M-Ras P40D in complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein M-Ras | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3KKN
 
 | | Crystal structure of H-Ras T35S in complex with GppNHp | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Muraoka, S, Shima, F, Liao, J, Ijiri, Y, Matsumoto, K, Ye, M, Inoue, T, Kataoka, T. | | Deposit date: | 2009-11-06 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis for conformational dynamics of GTP-bound Ras protein
J.Biol.Chem., 285, 2010
|
|
3LNK
 
 | | Structure of BACE bound to SCH743813 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-{(1S,2S)-1-(3,5-difluorobenzyl)-2-hydroxy-2-[(2R)-4-(phenylcarbonyl)piperazin-2-yl]ethyl}-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Orth, P, Cumming, J. | | Deposit date: | 2010-02-02 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3L5D
 
 | | Structure of BACE Bound to SCH723873 | | Descriptor: | 1-butyl-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5C
 
 | | Structure of BACE Bound to SCH723871 | | Descriptor: | 1-(4-cyanophenyl)-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3L5F
 
 | | Structure of BACE Bound to SCH736201 | | Descriptor: | (2E,5R)-5-(2-cyclohexylethyl)-5-(cyclohexylmethyl)-2-imino-3-methylimidazolidin-4-one, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
3LPJ
 
 | | Structure of BACE Bound to SCH743641 | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N'-[(1S,2S)-2-[(2R)-4-benzylpiperazin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2010-02-05 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Piperazine sulfonamide BACE1 inhibitors: design, synthesis, and in vivo characterization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7SWO
 
 | | C98C7 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | C98C7 Fab heavy chain, C98C7 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
7SWP
 
 | | G32Q4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | G32Q4 Fab heavy chain, G32Q4 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
7SWN
 
 | | G32A4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | G32A4 Fab heavy chain, G32A4 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
6VTI
 
 | | Solution NMR structure of the N-terminal domain of the Serine/threonine-protein phosphatase 1 regulatory subunit 10, PPP1R10 | | Descriptor: | Serine/threonine-protein phosphatase 1 regulatory subunit 10 | | Authors: | Lemak, A, Wei, Y, Duan, S, Houliston, S, Penn, L.Z, Arrowsmith, C.H. | | Deposit date: | 2020-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The MYC oncoprotein directly interacts with its chromatin cofactor PNUTS to recruit PP1 phosphatase.
Nucleic Acids Res., 50, 2022
|
|
7LQT
 
 | | Solution NMR structure of the PNUTS amino-terminal Domain fused to Myc Homology Box 0 | | Descriptor: | Serine/threonine-protein phosphatase 1 regulatory subunit 10,Myc proto-oncogene protein fusion | | Authors: | Lemak, A, Wei, Y, Duan, S, Houliston, S, Penn, L.Z, Arrowsmith, C.H. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The MYC oncoprotein directly interacts with its chromatin cofactor PNUTS to recruit PP1 phosphatase.
Nucleic Acids Res., 50, 2022
|
|
6VKG
 
 | | Human carbonic anhydrase IX mimic with Epacadostat bound | | Descriptor: | N-(3-bromo-4-fluorophenyl)-N'-hydroxy-4-{[2-(sulfamoylamino)ethyl]amino}-1,2,5-oxadiazole-3-carboximidamide, SULFATE ION, ZINC ION, ... | | Authors: | Peat, T.S, Carta, F, Supuran, C.T. | | Deposit date: | 2020-01-20 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | X-ray crystallography of Epacadostat in adduct with Carbonic Anhydrase IX.
Bioorg.Chem., 97, 2020
|
|
4WYQ
 
 | | Crystal structure of the Dicer-TRBP interface | | Descriptor: | Endoribonuclease Dicer, Poly(UNK), RISC-loading complex subunit TARBP2 | | Authors: | Wilson, R.C, Doudna, J.A. | | Deposit date: | 2014-11-18 | | Release date: | 2014-12-17 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dicer-TRBP Complex Formation Ensures Accurate Mammalian MicroRNA Biogenesis.
Mol.Cell, 57, 2015
|
|
2QMV
 
 | |
8IYP
 
 | | Crystal structure of serine palmitoyltransferase soaked in 190 mM D-serine solution | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal structure of Serine Palmitoyltransferase from Sphingobacterium multivorum
To Be Published
|
|
8IYT
 
 | | Crystal Structure of Serine Palmitoyltransferase complexed with D-methylserine | | Descriptor: | (2~{R})-2-methyl-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-3-oxidanyl-propanoic acid, 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Serine Palmitoyltransferase from Sphingobacterium multivorum
To Be Published
|
|
7TO4
 
 | | Structural and functional impact by SARS-CoV-2 Omicron spike mutations | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2022-01-22 | | Release date: | 2022-02-16 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional impact by SARS-CoV-2 Omicron spike mutations.
Cell Rep, 39, 2022
|
|
