5WQC
 
 | | Crystal structure of human orexin 2 receptor bound to the selective antagonist EMPA determined by the synchrotron light source at SPring-8. | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Hirata, K, Yamashita, K, Tsujimoto, H, Sasanuma, M, Horita, S, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-11-25 | | Release date: | 2017-11-29 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA
Structure, 26, 2018
|
|
1QTR
 
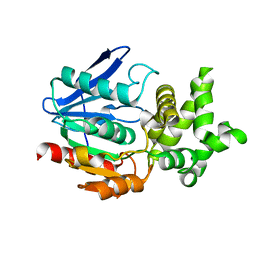 | | CRYSTAL STRUCTURE ANALYSIS OF THE PROLYL AMINOPEPTIDASE FROM SERRATIA MARCESCENS | | Descriptor: | PROLYL AMINOPEPTIDASE | | Authors: | Yoshimoto, T, Kabashima, T, Uchikawa, K, Inoue, T, Tanaka, N. | | Deposit date: | 1999-06-28 | | Release date: | 1999-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of prolyl aminopeptidase from Serratia marcescens.
J.Biochem.(Tokyo), 126, 1999
|
|
2N9L
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in-cell GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
2N9K
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for in vitro GB1 | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Ikeya, T, Hanashima, T, Hosoya, S, Shimazaki, M, Ikeda, S, Mishima, M, Guentert, P, Ito, Y. | | Deposit date: | 2015-11-26 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Improved in-cell structure determination of proteins at near-physiological concentration
Sci Rep, 6, 2016
|
|
8KAD
 
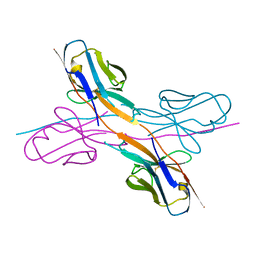 | | Crystal structure of an antibody light chain tetramer with 3D domain swapping | | Descriptor: | Antibody light chain | | Authors: | Sakai, T, Mashima, T, Kobayashi, N, Ogata, H, Uda, T, Hifumi, E, Hirota, S. | | Deposit date: | 2023-08-02 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic insights into antibody light chain tetramer formation through 3D domain swapping.
Nat Commun, 14, 2023
|
|
8IC7
 
 | | exo-beta-D-arabinofuranosidase ExoMA2 from Microbacterium arabinogalactanolyticum in complex with beta-D-arabinofuranose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Fukushima, R, Kashima, T, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8IC6
 
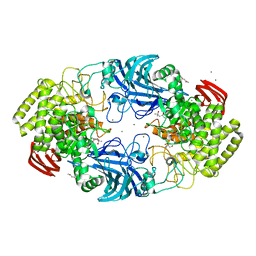 | | exo-beta-D-arabinanase ExoMA2 from Microbacterium arabinogalactanolyticum in complex with Tris | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Fukushima, R, Kashima, T, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
1BDJ
 
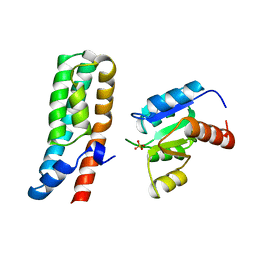 | | COMPLEX STRUCTURE OF HPT DOMAIN AND CHEY | | Descriptor: | AEROBIC RESPIRATION CONTROL SENSOR PROTEIN ARCB, CHEY, SULFATE ION | | Authors: | Kato, M, Mizuno, T, Shimizu, T, Hakoshima, T. | | Deposit date: | 1998-05-10 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure of the histidine-containing phosphotransfer (HPt) domain of the anaerobic sensor protein ArcB complexed with the chemotaxis response regulator CheY.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1NPM
 
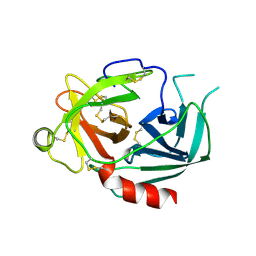 | | NEUROPSIN, A SERINE PROTEASE EXPRESSED IN THE LIMBIC SYSTEM OF MOUSE BRAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NEUROPSIN | | Authors: | Kishi, T, Kato, M, Shimizu, T, Kato, K, Matsumoto, K, Yoshida, S, Shiosaka, S, Hakoshima, T. | | Deposit date: | 1998-01-07 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of neuropsin, a hippocampal protease involved in kindling epileptogenesis.
J.Biol.Chem., 274, 1999
|
|
1DJ6
 
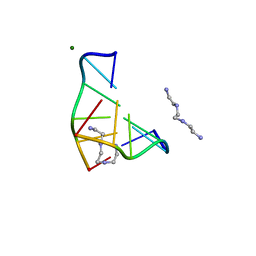 | | COMPLEX OF A Z-DNA HEXAMER, D(CG)3, WITH SYNTHETIC POLYAMINE AT ROOM TEMPERATURE | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', MAGNESIUM ION, N,N'-BIS(2-AMINOETHYL)-1,2-ETHANEDIAMINE | | Authors: | Ohishi, H, Tomita, K.-i, Nakanishi, I, Ohtsuchi, M, Hakoshima, T, Rich, A. | | Deposit date: | 1999-12-01 | | Release date: | 1999-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The crystal structure of N1-[2-(2-amino-ethylamino)-ethyl]-ethane-1,2-diamine (polyamines) binding to the minor groove of d(CGCGCG)2, hexamer at room temperature
FEBS Lett., 523, 2002
|
|
5D4H
 
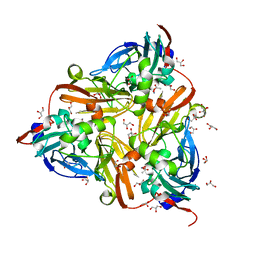 | | High-resolution nitrite complex of a copper nitrite reductase determined by synchrotron radiation crystallography | | Descriptor: | ACETIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D4I
 
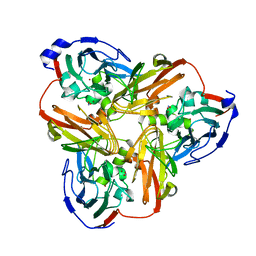 | | Intact nitrite complex of a copper nitrite reductase determined by serial femtosecond crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2A0B
 
 | | HISTIDINE-CONTAINING PHOSPHOTRANSFER DOMAIN OF ARCB FROM ESCHERICHIA COLI | | Descriptor: | HPT DOMAIN, ZINC ION | | Authors: | Kato, M, Mizuno, T, Shimizu, T, Hakoshima, T. | | Deposit date: | 1998-04-02 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Refined structure of the histidine-containing phosphotransfer (HPt) domain of the anaerobic sensor kinase ArcB from Escherichia coli at 1.57 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
8GR9
 
 | | Crystal structure of peroxisomal citrate synthase (Cit2) from Saccharomyces cerevisiae in complex with oxaloacetate and coenzyme-A | | Descriptor: | CHLORIDE ION, COENZYME A, Citrate synthase, ... | | Authors: | Nishio, K, Nakatsukasa, K, Kamura, T, Mizushima, T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress.
Sci Adv, 9, 2023
|
|
8GR8
 
 | | Crystal structure of peroxisomal citrate synthase (Cit2) from Saccharomycescerevisiae | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Citrate synthase, ... | | Authors: | Nishio, K, Nakatsukasa, K, Kamura, T, Mizushima, T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress.
Sci Adv, 9, 2023
|
|
8GRE
 
 | | F-box protein in complex with skp1(FL) and substrate | | Descriptor: | Citrate synthase, E3 ubiquitin ligase complex SCF subunit, F-box protein UCC1, ... | | Authors: | Nishio, K, Nakatsukasa, K, Kamura, T, Mizushima, T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress.
Sci Adv, 9, 2023
|
|
8GQZ
 
 | | Crystal structure of mitochondrial citrate synthase (Cit1) from Saccharomyces cerevisiae | | Descriptor: | ACETATE ION, CHLORIDE ION, Citrate synthase, ... | | Authors: | Nishio, K, Nakatsukasa, K, Kamura, T, Mizushima, T. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress.
Sci Adv, 9, 2023
|
|
8GRF
 
 | | Crystal structure of F-box protein in the ternary complex with adaptor protein Skp1(DL) and its substrate | | Descriptor: | 1,2-ETHANEDIOL, Citrate synthase, E3 ubiquitin ligase complex SCF subunit, ... | | Authors: | Nishio, K, Nakatsukasa, K, Kamura, T, Mizushima, T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Defective import of mitochondrial metabolic enzyme elicits ectopic metabolic stress.
Sci Adv, 9, 2023
|
|
3RKS
 
 | | Crystal Structure of the Manihot esculenta Hydroxynitrile Lyase (MeHNL) K176P mutant | | Descriptor: | GLYCEROL, Hydroxynitrilase | | Authors: | Cielo, C.B.C, Yamane, T, Asano, Y, Dadashipour, M, Suzuki, A, Mizushima, T, Komeda, H. | | Deposit date: | 2011-04-18 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic Studies of Manihot esculenta hydroxynitrile lyase Lysine-to-Proline mutants
To be Published
|
|
5F7A
 
 | | Nitrite complex structure of copper nitrite reductase from Alcaligenes faecalis determined at 293 K | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1GE9
 
 | | SOLUTION STRUCTURE OF THE RIBOSOME RECYCLING FACTOR | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Yoshida, T, Uchiyama, S, Nakano, H, Kashimori, H, Kijima, H, Ohshima, T, Saihara, Y, Ishino, T, Shimahara, T, Yoshida, T, Yokose, K, Ohkubo, T, Kaji, A, Kobayashi, Y. | | Deposit date: | 2000-10-19 | | Release date: | 2001-05-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosome recycling factor from Aquifex aeolicus.
Biochemistry, 40, 2001
|
|
5F7B
 
 | | Resting state structure of CuNiR form Alcaligenes faecalis determined at 293 K | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2D10
 
 | |
2D11
 
 | |
7F76
 
 | |
