7ZSU
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, Steroid C26-monooxygenase, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-05-09 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7ZLZ
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, Steroid C26-monooxygenase, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-04-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
5A35
 
 | | Crystal structure of Glycine Cleavage Protein H-Like (GcvH-L) from Streptococcus pyogenes | | Descriptor: | GLYCINE CLEAVAGE SYSTEM H PROTEIN, PENTAETHYLENE GLYCOL | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
3SIJ
 
 | |
3SII
 
 | |
5A3A
 
 | | Crystal structure of the ADP-ribosylating sirtuin (SirTM) from Streptococcus pyogenes (Apo form) | | Descriptor: | 1,2-ETHANEDIOL, GLYCINE, SIR2 FAMILY PROTEIN, ... | | Authors: | Rack, J.G.M, Morra, R, Barkauskaite, E, Kraehenbuehl, R, Ariza, A, Qu, Y, Ortmayer, M, Leidecker, O, Cameron, D.R, Matic, I, Peleg, A.Y, Leys, D, Traven, A, Ahel, I. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of a Class of Protein Adp-Ribosylating Sirtuins in Microbial Pathogens.
Mol.Cell, 59, 2015
|
|
5D6S
 
 | | Structure of epoxyqueuosine reductase from Streptococcus thermophilus. | | Descriptor: | COBALAMIN, Epoxyqueuosine reductase, IRON/SULFUR CLUSTER | | Authors: | Payne, K.A.P, Fisher, K, Dunstan, M.S, Sjuts, H, Leys, D. | | Deposit date: | 2015-08-12 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Epoxyqueuosine Reductase Structure Suggests a Mechanism for Cobalamin-dependent tRNA Modification.
J.Biol.Chem., 290, 2015
|
|
3SIH
 
 | |
5E1W
 
 | |
5E20
 
 | | Crystal structure of the organohalide sensing RdhR-CbdbA1625 transcriptional regulator in the 2,3-dichlorophenol bound form | | Descriptor: | 2,3-dichlorophenol, Transcriptional regulator, MarR family | | Authors: | Quezada, C.P, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structures of RdhRCbdbA1625 provide insight into sensing of chloroaromatic compounds by MarR-type regulators
To be published
|
|
5E1Z
 
 | | Crystal structure of the organohalide sensing RdhR-CbdbA1625 transcriptional regulator in the 2,4-dichlorophenol bound form | | Descriptor: | 2,4-dichlorophenol, Transcriptional regulator, MarR family | | Authors: | Quezada, C.P, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structures of RdhRCbdbA1625 provide insight into sensing of chloroaromatic compounds by MarR-type regulators
to be published
|
|
5E1X
 
 | | Crystal structure of the organohalide sensing RdhR-CbdbA1625 transcriptional regulator in the 3,4-dichlorophenol bound form | | Descriptor: | 3,4-dichlorophenol, Transcriptional regulator, MarR family | | Authors: | Quezada, C.P, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal structures of RdhRCbdbA1625 provide insight into sensing of chloroaromatic compounds by MarR-type regulators
TO BE PUBLISHED
|
|
1CXY
 
 | | STRUCTURE AND CHARACTERIZATION OF ECTOTHIORHODOSPIRA VACUOLATA CYTOCHROME B558, A PROKARYOTIC HOMOLOGUE OF CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kostanjevecki, V, Leys, D, Van Driessche, G, Meyer, T.E, Cusanovich, M.A, Fischer, U, Guisez, Y, Van Beeumen, J. | | Deposit date: | 1999-08-31 | | Release date: | 1999-09-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and characterization of Ectothiorhodospira vacuolata cytochrome b(558), a prokaryotic homologue of cytochrome b(5).
J.Biol.Chem., 274, 1999
|
|
4KVQ
 
 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase wild type with palmitic acid bound | | Descriptor: | Aldehyde decarbonylase, FE (III) ION, PALMITIC ACID | | Authors: | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
1JRY
 
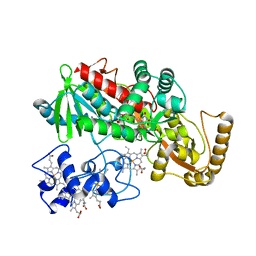 | | Crystal structure of Arg402Lys mutant flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C, FUMARIC ACID, ... | | Authors: | Mowat, C.G, Moysey, R, Miles, C.S, Leys, D, Doherty, M.K, Taylor, P, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2001-08-15 | | Release date: | 2001-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and crystallographic analysis of the key active site acid/base arginine in a soluble fumarate reductase.
Biochemistry, 40, 2001
|
|
8J2X
 
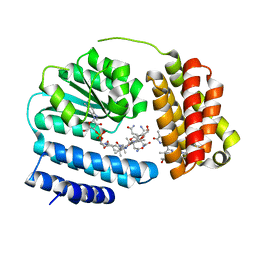 | | Saccharothrix syringae photocobilins protein, light state | | Descriptor: | BILIVERDINE IX ALPHA, COBALAMIN, Cobalamin-binding protein, ... | | Authors: | Zhang, S, Poddar, H, Levy, C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
8J2Y
 
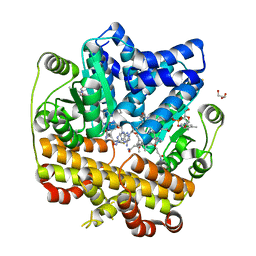 | | Acidimicrobiaceae bacterium photocobilins protein, dark state | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, S, Poddar, H, Levy, W.C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
8J2W
 
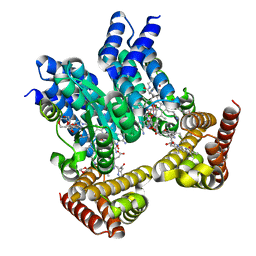 | | Saccharothrix syringae photocobilins protein, dark state | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5'-DEOXYADENOSINE, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, S, Poddar, H, Levy, W.C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
1KSU
 
 | | Crystal Structure of His505Tyr Mutant Flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pankhurst, K.L, Mowat, C.G, Miles, C.S, Leys, D, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2002-01-14 | | Release date: | 2002-08-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of His505 in the soluble fumarate reductase from Shewanella frigidimarina.
Biochemistry, 41, 2002
|
|
1JRX
 
 | | Crystal structure of Arg402Ala mutant flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C, FUMARIC ACID, ... | | Authors: | Mowat, C.G, Moysey, R, Miles, C.S, Leys, D, Doherty, M.K, Taylor, P, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2001-08-15 | | Release date: | 2001-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and crystallographic analysis of the key active site acid/base arginine in a soluble fumarate reductase.
Biochemistry, 40, 2001
|
|
1KSS
 
 | | Crystal Structure of His505Ala Mutant Flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pankhurst, K.L, Mowat, C.G, Miles, C.S, Leys, D, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2002-01-14 | | Release date: | 2002-08-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of His505 in the soluble fumarate reductase from Shewanella frigidimarina.
Biochemistry, 41, 2002
|
|
1JRZ
 
 | | Crystal structure of Arg402Tyr mutant flavocytochrome c3 from Shewanella frigidimarina | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C, FUMARIC ACID, ... | | Authors: | Mowat, C.G, Moysey, R, Miles, C.S, Leys, D, Doherty, M.K, Taylor, P, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2001-08-15 | | Release date: | 2001-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and crystallographic analysis of the key active site acid/base arginine in a soluble fumarate reductase.
Biochemistry, 40, 2001
|
|
7AE7
 
 | |
7AE4
 
 | |
7AE5
 
 | |
