3LUP
 
 | | Crystal structure of fatty acid binding DegV family protein SAG1342 from Streptococcus agalactiae | | Descriptor: | 9-OCTADECENOIC ACID, DegV family protein, GLYCEROL | | Authors: | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-18 | | Release date: | 2010-03-02 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of fatty acid binding DegV family protein SAG1342 from Streptococcus agalactiae
To be Published
|
|
3LUQ
 
 | | The Crystal Structure of a PAS Domain from a Sensory Box Histidine Kinase Regulator from Geobacter sulfurreducens to 2.5A | | Descriptor: | SULFATE ION, Sensor protein, TRIETHYLENE GLYCOL | | Authors: | Stein, A.J, Weger, A, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-18 | | Release date: | 2010-03-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Crystal Structure of a PAS Domain from a Sensory Box Histidine Kinase Regulator from Geobacter sulfurreducens to 2.5A
To be Published
|
|
3LVU
 
 | | Crystal structure of ABC transporter, periplasmic substrate-binding protein SPO2066 from Silicibacter pomeroyi | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ABC transporter, ... | | Authors: | Chang, C, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of ABC transporter, periplasmic substrate-binding protein SPO2066 from Silicibacter pomeroyi
To be Published
|
|
3LXQ
 
 | | The Crystal Structure of a Protein in the Alkaline Phosphatase Superfamily from Vibrio parahaemolyticus to 1.95A | | Descriptor: | CHLORIDE ION, Uncharacterized protein VP1736 | | Authors: | Stein, A.J, Weger, A, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of a Protein in the Alkaline Phosphatase Superfamily from Vibrio parahaemolyticus to 1.95A
To be Published
|
|
3M4R
 
 | | Structure of the N-terminal Class II Aldolase domain of a conserved protein from Thermoplasma acidophilum | | Descriptor: | CHLORIDE ION, Uncharacterized protein, ZINC ION | | Authors: | Cuff, M.E, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-11 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the N-terminal Class II Aldolase domain of a conserved protein from Thermoplasma acidophilum
TO BE PUBLISHED
|
|
2G7Z
 
 | | Conserved DegV-like Protein of Unknown Function from Streptococcus pyogenes M1 GAS Binds Long-chain Fatty Acids | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, ... | | Authors: | Nocek, B, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-01 | | Release date: | 2006-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conserved hypothetical protein from Streptococcus pyogenes M1 GAS discloses long-fatty acid (heptadecanoic acid) binding function
To be Published
|
|
3M0Z
 
 | | Crystal structure of putative aldolase from Klebsiella pneumoniae. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Chang, C, Rakowski, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of putative aldolase from Klebsiella pneumoniae.
To be Published
|
|
2G2C
 
 | |
6GT9
 
 | | Crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with galactose | | Descriptor: | Putative sugar binding protein, SULFATE ION, beta-D-galactopyranose | | Authors: | Sherf, D, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2018-06-16 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | The crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus, in complex with galactose
To Be Published
|
|
4URK
 
 | | PI3Kg in complex with AZD6482 | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Giordanetto, F, Barlaam, B, Berglund, S, Edman, K, Karlsson, O, Lindberg, J, Nylander, S, Inghardt, T. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of 9-(1-Phenoxyethyl)-2-Morpholino-4-Oxo-Pyrido[1, 2-A]Pyrimidine-7-Carboxamides as Oral Pi3Kbeta Inhibitors, Useful as Antiplatelet Agents.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4V96
 
 | | The structure of a 1.8 MDa viral genome injection device suggests alternative infection mechanisms | | Descriptor: | BPP, ORF46, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V, van Sinderen, D, Cambillau, C. | | Deposit date: | 2012-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1ZRU
 
 | | structure of the lactophage p2 receptor binding protein in complex with glycerol | | Descriptor: | GLYCEROL, lactophage p2 receptor binding protein | | Authors: | Spinelli, S, Tremblay, D.M, Tegoni, M, Blangy, S, Huyghe, C, Desmyter, A, Labrie, S, de Haard, H, Moineau, S, Cambillau, C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-22 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site.
J.Bacteriol., 188, 2006
|
|
4UQX
 
 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, HSIE1 | | Authors: | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
J.Biol.Chem., 289, 2014
|
|
4UQW
 
 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | Descriptor: | BENZAMIDINE, PROTEIN CLPV1 | | Authors: | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
J.Biol.Chem., 289, 2014
|
|
4W66
 
 | |
4UQY
 
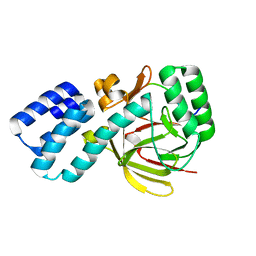 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | Descriptor: | HSIB1, HSIE1 | | Authors: | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
J.Biol.Chem., 289, 2014
|
|
4WER
 
 | | Crystal structure of diacylglycerol kinase catalytic domain protein from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Diacylglycerol kinase catalytic domain protein | | Authors: | Chang, C, Clancy, S, Hatzos-Skintges, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-10 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of diacylglycerol kinase catalytic domain protein from Enterococcus faecalis V583
To Be Published
|
|
8FU9
 
 | | Structure of Covid Spike variant deltaN25 with one erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
8FU7
 
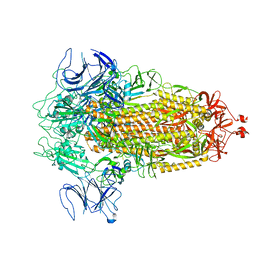 | | Structure of Covid Spike variant deltaN135 in fully closed form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
8FU8
 
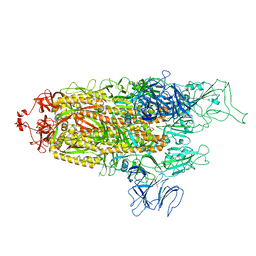 | | Structure of Covid Spike variant deltaN135 with one erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yu, X, Juraszek, J, Rutten, L, Bakkers, M.J.G, Blokland, S, Van den Broek, N.J.F, Verwilligen, A.Y.W, Abeywickrema, P, Vingerhoets, J, Neefs, J, Bakhash, S.A.M, Roychoudhury, P, Greninger, A, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-01-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Convergence of immune escape strategies highlights plasticity of SARS-CoV-2 spike.
Plos Pathog., 19, 2023
|
|
6OBZ
 
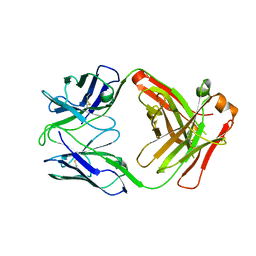 | | Crystal structure of FluA-20 Fab | | Descriptor: | Heavy chain of FluA-20 Fab, Light chain of FluA-20 Fab, Immunoglobulin light chain | | Authors: | Wilson, I.A, Lang, S. | | Deposit date: | 2019-03-21 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface.
Cell, 177, 2019
|
|
6OCB
 
 | |
6OC3
 
 | |
1K1G
 
 | | STRUCTURAL BASIS FOR RECOGNITION OF THE INTRON BRANCH SITE RNA BY SPLICING FACTOR 1 | | Descriptor: | 5'-R(*UP*AP*UP*AP*CP*UP*AP*AP*CP*AP*A)-3', SF1-Bo isoform | | Authors: | Liu, Z, Luyten, I, Bottomley, M.J, Messias, A.C, Houngninou-Molango, S, Sprangers, R, Zanier, K, Kramer, A, Sattler, M. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of the intron branch site RNA by splicing factor 1.
Science, 294, 2001
|
|
6YRV
 
 | | Crystal structure of FAP after illumination at 100K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
