7EL1
 
 | | Structure of a protein from bacteria | | Descriptor: | 100AA, CRISPR-associated endonuclease Cas9, DNA (28-MER), ... | | Authors: | Liu, H, Zhu, Y, Huang, Z. | | Deposit date: | 2021-04-07 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis of Staphylococcus aureus Cas9 inhibition by AcrIIA14.
Nucleic Acids Res., 49, 2021
|
|
7EPQ
 
 | |
7FAH
 
 | | Immune complex of head region of CA09 HA and neutralizing antibody 12H5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Li, T.T, Xue, W.H, Gu, Y, Li, S.W. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
7WMD
 
 | | PQQ-dependent alcohol dehydrogenase detoxifying DON | | Descriptor: | CALCIUM ION, PQQ-dependent alcohol dehydrogenase | | Authors: | Chen, M, Yang, H, Lv, F. | | Deposit date: | 2022-01-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Function Analysis of a Quinone-Dependent Dehydrogenase Capable of Deoxynivalenol Detoxification.
J.Agric.Food Chem., 70, 2022
|
|
7WMK
 
 | | PQQ-dependent alcohol dehydrogenase complexed with PQQ | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Chen, M, Yang, H, Lv, F. | | Deposit date: | 2022-01-15 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Function Analysis of a Quinone-Dependent Dehydrogenase Capable of Deoxynivalenol Detoxification.
J.Agric.Food Chem., 70, 2022
|
|
6AGY
 
 | | Aspergillus fumigatus Af293 NDK | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Hu, Y, Han, L. | | Deposit date: | 2018-08-15 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of crystal structure and key residues of Aspergillus fumigatus nucleoside diphosphate kinase.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
7WLP
 
 | |
7W7P
 
 | |
7YHK
 
 | | Cryo-EM structure of the HA trimer of A/Beijing/262/1995(H1N1) in complex with neutralizing antibody 12H5 | | Descriptor: | 12H5 heavy chain, 12H5 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, Q, Li, S, Li, T, Xue, W, Sun, H. | | Deposit date: | 2022-07-13 | | Release date: | 2022-08-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
1UBS
 
 | | TRYPTOPHAN SYNTHASE (E.C.4.2.1.20) WITH A MUTATION OF LYS 87->THR IN THE B SUBUNIT AND IN THE PRESENCE OF LIGAND L-SERINE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE, SODIUM ION, ... | | Authors: | Rhee, S, Parris, K, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1995-12-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
1WUT
 
 | | Acyl Ureas as Human Liver Glycogen Phosphorylase Inhibitors for the Treatment of Type 2 Diabetes | | Descriptor: | 7-[2,6-DICHLORO-4-({[(2-CHLOROBENZOYL)AMINO]CARBONYL}AMINO)PHENOXY]HEPTANOIC ACID, Glycogen phosphorylase, muscle form, ... | | Authors: | Klabunde, T, Wendt, K.U, Kadereit, D, Brachvogel, V, Burger, H.-J, Herling, A.W, Oikonomakos, N.G, Kosmopoulou, M.N, Schmoll, D, Sarubbi, E, von Roedern, E, Schonafinger, K, Defossa, E. | | Deposit date: | 2004-12-08 | | Release date: | 2005-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystallographic studies on acyl ureas, a new class of glycogen phosphorylase inhibitors, as potential antidiabetic drugs
Protein Sci., 14, 2005
|
|
8H66
 
 | |
8H6D
 
 | |
8H6C
 
 | |
8H65
 
 | |
8HY0
 
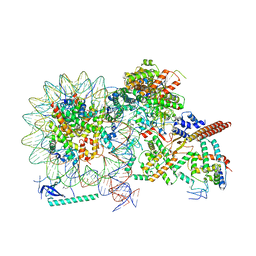 | |
8HXZ
 
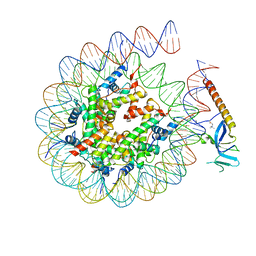 | | Cryo-EM structure of Eaf3 CHD in complex with nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (352-MER), Histone H2A, ... | | Authors: | Cui, H, Wang, H. | | Deposit date: | 2023-01-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of histone deacetylase complex Rpd3S bound to nucleosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8HXX
 
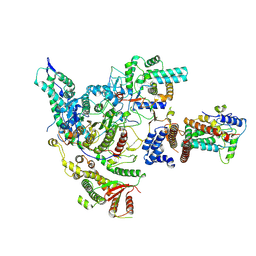 | | Cryo-EM structure of the histone deacetylase complex Rpd3S | | Descriptor: | Chromatin modification-related protein EAF3, Histone H3, Histone deacetylase RPD3, ... | | Authors: | Cui, H, Wang, H. | | Deposit date: | 2023-01-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of histone deacetylase complex Rpd3S bound to nucleosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8HXY
 
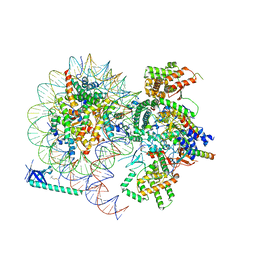 | |
8JHO
 
 | |
7ZOI
 
 | | Carbohydrate binding domain CBM92-A from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 | | Descriptor: | Glycoside hydrolase family 18 | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOO
 
 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with gentiobiose | | Descriptor: | Glycoside hydrolase family 18, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZON
 
 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with glucose | | Descriptor: | Glycoside hydrolase family 18, PENTAETHYLENE GLYCOL, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOH
 
 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 | | Descriptor: | Glycoside hydrolase family 18 | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOP
 
 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with sophorose. | | Descriptor: | Glycoside hydrolase family 18, beta-D-glucopyranose | | Authors: | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
