7X4Q
 
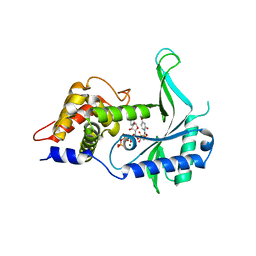 | | LpCdnE UTP Mg complex | | Descriptor: | Cyclic dipyrimidine nucleotide synthase, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4G
 
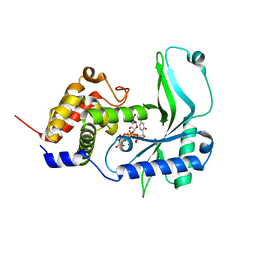 | | CD-NTase ClCdnE in complex with substrate UTP | | Descriptor: | ClCdnE, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4T
 
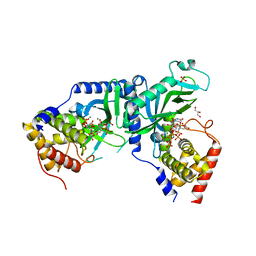 | | LpCdnE UMPNPP Mg complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, Cyclic dipyrimidine nucleotide synthase, GLYCEROL, ... | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
2AFX
 
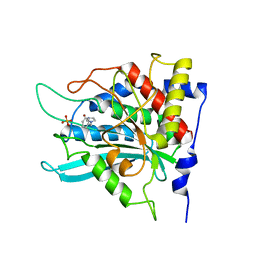 | | Crystal structure of human glutaminyl cyclase in complex with 1-benzylimidazole | | Descriptor: | 1-BENZYL-1H-IMIDAZOLE, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AFS
 
 | | Crystal structure of the genetic mutant R54W of human glutaminyl cyclase | | Descriptor: | Glutaminyl-peptide cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AFZ
 
 | | Crystal structure of human glutaminyl cyclase in complex with 1-vinylimidazole | | Descriptor: | 1-VINYLIMIDAZOLE, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AZJ
 
 | | Crystal structure for the mutant D81C of Sulfolobus solfataricus hexaprenyl pyrophosphate synthase | | Descriptor: | Geranylgeranyl pyrophosphate synthetase | | Authors: | Sun, H.Y, Ko, T.P, Kuo, C.J, Guo, R.T, Chou, C.C, Liang, P.H, Wang, A.H.J. | | Deposit date: | 2005-09-11 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Homodimeric hexaprenyl pyrophosphate synthase from the thermoacidophilic crenarchaeon Sulfolobus solfataricus displays asymmetric subunit structures
J.Bacteriol., 187, 2005
|
|
2AFM
 
 | | Crystal structure of human glutaminyl cyclase at pH 6.5 | | Descriptor: | Glutaminyl-peptide cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AFO
 
 | | Crystal structure of human glutaminyl cyclase at pH 8.0 | | Descriptor: | Glutaminyl-peptide cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AFW
 
 | | Crystal structure of human glutaminyl cyclase in complex with N-acetylhistamine | | Descriptor: | Glutaminyl-peptide cyclotransferase, N-[2-(1H-IMIDAZOL-4-YL)ETHYL]ACETAMIDE, SULFATE ION, ... | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AFU
 
 | | Crystal structure of human glutaminyl cyclase in complex with glutamine t-butyl ester | | Descriptor: | Glutaminyl-peptide cyclotransferase, TERT-BUTYL D-ALPHA-GLUTAMINATE, ZINC ION | | Authors: | Huang, K.F, Liu, Y.L, Cheng, W.J, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2005-07-26 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structures of human glutaminyl cyclase, an enzyme responsible for protein N-terminal pyroglutamate formation
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AZK
 
 | | Crystal structure for the mutant W136E of Sulfolobus solfataricus hexaprenyl pyrophosphate synthase | | Descriptor: | Geranylgeranyl pyrophosphate synthetase | | Authors: | Sun, H.Y, Ko, T.P, Kuo, C.J, Guo, R.T, Chou, C.C, Liang, P.H, Wang, A.H.J. | | Deposit date: | 2005-09-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Homodimeric hexaprenyl pyrophosphate synthase from the thermoacidophilic crenarchaeon Sulfolobus solfataricus displays asymmetric subunit structures
J.Bacteriol., 187, 2005
|
|
2AZL
 
 | | Crystal structure for the mutant F117E of Thermotoga maritima octaprenyl pyrophosphate synthase | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Sun, H.Y, Ko, T.P, Kuo, C.J, Guo, R.T, Chou, C.C, Liang, P.H, Wang, A.H. | | Deposit date: | 2005-09-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Homodimeric hexaprenyl pyrophosphate synthase from the thermoacidophilic crenarchaeon Sulfolobus solfataricus displays asymmetric subunit structures
J.Bacteriol., 187, 2005
|
|
4QNN
 
 | |
1VG2
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima A76Y mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
1VG6
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima F132A/L128A/I123A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
1VG7
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima F132A/L128A/I123A/D62A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
1VG3
 
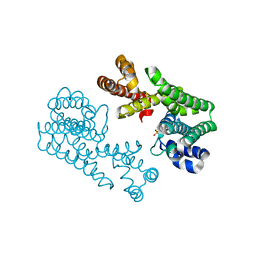 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima A76Y/S77F mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
1VG4
 
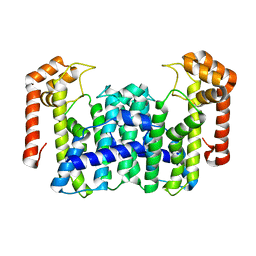 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima F132A/L128A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
5ITJ
 
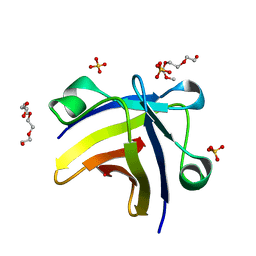 | | The structure of histone-like protein | | Descriptor: | AbrB family transcriptional regulator, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Lin, B.L, Chen, C.Y, Huang, C.H, Ko, T.P, Chiang, C.H, Lin, K.F, Chang, Y.C, Lin, P.Y, Tsai, H.H.G, Wang, A.H.J. | | Deposit date: | 2016-03-17 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The Arginine Pairs and C-Termini of the Sso7c4 from Sulfolobus solfataricus Participate in Binding and Bending DNA.
PLoS ONE, 12, 2017
|
|
5ITM
 
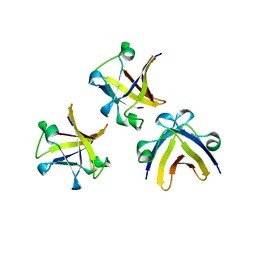 | | The structure of truncated histone-like protein | | Descriptor: | AbrB family transcriptional regulator | | Authors: | Lin, B.L, Chen, C.Y, Huang, C.H, Ko, T.P, Chiang, C.H, Lin, K.F, Chang, Y.C, Lin, P.Y, Tsai, H.H.G, Wang, A.H.J. | | Deposit date: | 2016-03-17 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Arginine Pairs and C-Termini of the Sso7c4 from Sulfolobus solfataricus Participate in Binding and Bending DNA.
PLoS ONE, 12, 2017
|
|
5H9D
 
 | | Crystal structure of Heptaprenyl Diphosphate Synthase from Staphylococcus aureus | | Descriptor: | C-terminal peptide from Heptaprenyl diphosphate synthase (HEPPP synthase) subunit 1 family protein, Farnesyl pyrophosphate synthetase, Heptaprenyl diphosphate synthase (HEPPP synthase) subunit 1 family protein, ... | | Authors: | Wei, H.L, Liu, W.D, Zheng, Y.Y, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-12-28 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure, Function, and Inhibition of Staphylococcus aureus Heptaprenyl Diphosphate Synthase
ChemMedChem, 11, 2016
|
|
6JD1
 
 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) in complex with Mg2+, NADH, and CPD at pH7.5 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, Putative ketol-acid reductoisomerase 2, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
6JD2
 
 | | Crystal structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) in complex with Mg2+ at pH8.5 | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Putative ketol-acid reductoisomerase 2 | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J.Am.Chem.Soc., 141, 2019
|
|
6JCW
 
 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) with Mg2+ at pH8.5 | | Descriptor: | MAGNESIUM ION, ketol-acid reductoisomerase | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
