3VRV
 
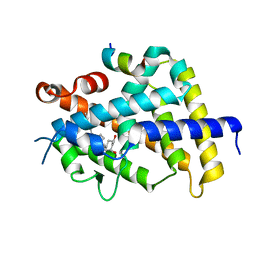 | | VDR ligand binding domain in complex with 2-Methylidene-26,27-dimethyl-19,24-dinor-1alpha,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R)-5-ethyl-5-hydroxyheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Yoshimoto, N, Inaba, Y, Itoh, T, Nakabayashi, M, Ito, N, Yamamoto, K. | | Deposit date: | 2012-04-14 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Butyl pocket formation in the vitamin d receptor strongly affects the agonistic or antagonistic behavior of ligands
J.Med.Chem., 55, 2012
|
|
3WC7
 
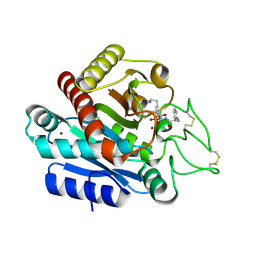 | | Carboxypeptidase B in complex with EF6265 | | Descriptor: | (2S)-7-amino-2-{[(R)-hydroxy{(1R)-2-methyl-1-[(3-phenylpropanoyl)amino]propyl}phosphoryl]methyl}heptanoic acid, Carboxypeptidase B, ZINC ION | | Authors: | Yoshimoto, N, Itoh, T, Inaba, Y, Yamamoto, K. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of carboxypeptidase B by selenium-containing inhibitor: selenium coordinates to zinc in enzyme.
J.Med.Chem., 56, 2013
|
|
3VLW
 
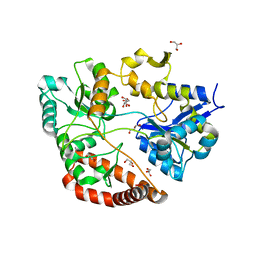 | | Crystal structure of Sphingomonas sp. A1 alginate-binding protein AlgQ1 in complex with mannuronate-guluronate disaccharide | | Descriptor: | AlgQ1, CALCIUM ION, GLYCEROL, ... | | Authors: | Nishitani, Y, Maruyama, Y, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter
Biochemistry, 51, 2012
|
|
3VRT
 
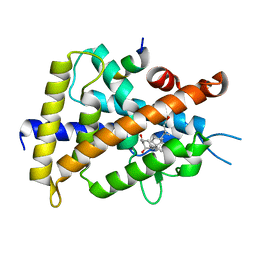 | | VDR ligand binding domain in complex with 2-Mehylidene-19,25,26,27-tetranor-1alpha,24-dihydroxyvitaminD3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R)-5-hydroxypentan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Yoshimoto, N, Inaba, Y, Itoh, T, Ito, N, Yamamoto, K. | | Deposit date: | 2012-04-14 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Butyl pocket formation in the vitamin d receptor strongly affects the agonistic or antagonistic behavior of ligands
J.Med.Chem., 55, 2012
|
|
3VW4
 
 | | Crystal structure of the DNA-binding domain of ColE2-P9 Rep in complex with the replication origin | | Descriptor: | DNA (5'-D(P*AP*AP*TP*GP*AP*GP*AP*CP*CP*AP*GP*AP*TP*AP*AP*GP*CP*CP*TP*TP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*AP*AP*GP*GP*CP*TP*TP*AP*TP*CP*TP*GP*GP*TP*CP*TP*CP*AP*TP*T)-3'), Rep, ... | | Authors: | Itou, H, Yagura, M, Itoh, T, Shirakihara, Y. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Replication Origin Unwinding by An Initiator-Primase of Plasmid ColE2-P9: Duplex DNA Unwinding by A Single Protein
J.Biol.Chem., 290, 2015
|
|
3WC5
 
 | | Carboxypeptidase B in complex with DD9 | | Descriptor: | (2R)-7-amino-2-(selanylmethyl)heptanoic acid, CACODYLATE ION, Carboxypeptidase B, ... | | Authors: | Yoshimoto, N, Itoh, T, Inaba, Y, Yamamoto, K. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-02 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for inhibition of carboxypeptidase B by selenium-containing inhibitor: selenium coordinates to zinc in enzyme.
J.Med.Chem., 56, 2013
|
|
3WT5
 
 | | A mixed population of antagonist and agonist binding conformers in a single crystal explains partial agonism against vitamin D receptor: Active vitamin D analogues with 22R-alkyl group | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3R)-3-butyl-6-hydroxy-6-methylheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-04-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Mixed Population of Antagonist and Agonist Binding Conformers in a Single Crystal Explains Partial Agonism against Vitamin D Receptor: Active Vitamin D Analogues with 22R-Alkyl Group.
J.Med.Chem., 57, 2014
|
|
3WT7
 
 | | Crystal structure of VDR-LBD complexed with 22R-Butyl-2-methylidene-26,27-dimethyl-19,24-dinor-1 ,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3R)-3-butyl-5-ethyl-5-hydroxyheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-04-08 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Mixed Population of Antagonist and Agonist Binding Conformers in a Single Crystal Explains Partial Agonism against Vitamin D Receptor: Active Vitamin D Analogues with 22R-Alkyl Group.
J.Med.Chem., 57, 2014
|
|
3WT6
 
 | | A mixed population of antagonist and agonist binding conformers in a single crystal explains partial agonism against vitamin D receptor: Active vitamin D analogues with 22R-alkyl group | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3R)-3-butyl-6-hydroxy-6-methylheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-04-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mixed Population of Antagonist and Agonist Binding Conformers in a Single Crystal Explains Partial Agonism against Vitamin D Receptor: Active Vitamin D Analogues with 22R-Alkyl Group.
J.Med.Chem., 57, 2014
|
|
3WC6
 
 | | Carboxypeptidase B in complex with 2nd zinc | | Descriptor: | ACETATE ION, Carboxypeptidase B, ZINC ION | | Authors: | Yoshimoto, N, Itoh, T, Inaba, Y, Yamamoto, K. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for inhibition of carboxypeptidase B by selenium-containing inhibitor: selenium coordinates to zinc in enzyme.
J.Med.Chem., 56, 2013
|
|
3X1H
 
 | | hPPARgamma Ligand binding domain in complex with 5-oxo-tricosahexaenoic acid | | Descriptor: | (7E,11Z,14Z,17Z,20Z)-5-oxotricosa-7,11,14,17,20-pentaenoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-11-18 | | Release date: | 2015-04-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of covalent bond formation between PPAR gamma and oxo-fatty acids.
Bioconjug.Chem., 26, 2015
|
|
3X1I
 
 | | hPPARgamma Ligand binding domain in complex with 6-oxo-tetracosahexaenoic acid | | Descriptor: | (8E,12Z,15Z,18Z,21Z)-6-oxotetracosa-8,12,15,18,21-pentaenoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-11-18 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of covalent bond formation between PPAR gamma and oxo-fatty acids.
Bioconjug.Chem., 26, 2015
|
|
2MLO
 
 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a Membrane bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
2MLQ
 
 | | Human CCR2 Membrane-Proximal C-Terminal Region (PRO-C) in a frount bound form | | Descriptor: | MCP-1 receptor | | Authors: | Esaki, K, Yoshinaga, S, Tsuji, T, Toda, E, Terashima, Y, Saitoh, T, Kohda, D, Kohno, T, Osawa, M, Ueda, T, Shimada, I, Matsushima, K, Terasawa, H. | | Deposit date: | 2014-03-04 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the binding of the membrane-proximal C-terminal region of chemokine receptor CCR2 with the cytosolic regulator FROUNT.
Febs J., 281, 2014
|
|
5XOF
 
 | | Crystal structure of human paired immunoglobulin-like type 2 receptor alpha with synthesized glycopeptide I | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Paired immunoglobulin-like type 2 receptor alpha, Peptide from Nitric oxide synthase, ... | | Authors: | Furukawa, A, Kakita, K, Yamada, T, Ishizuka, M, Sakamoto, J, Hatori, N, Maeda, N, Ohsaka, F, Saitoh, T, Nomura, T, Kuroki, K, Nambu, H, Arase, H, Matsunaga, H, Anada, M, Ose, T, Hashimoto, S, Maenaka, K. | | Deposit date: | 2017-05-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.963 Å) | | Cite: | Structural and thermodynamic analyses reveal critical features of glycopeptide recognition by the human PILR alpha immune cell receptor.
J. Biol. Chem., 292, 2017
|
|
5XO2
 
 | | Crystal structure of human paired immunoglobulin-like type 2 receptor alpha with synthesized glycopeptide II | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-6)-2-acetamido-2,4-dideoxy-alpha-D-xylo-hexopyranose, Paired immunoglobulin-like type 2 receptor alpha, Peptide from Envelope glycoprotein B | | Authors: | Furukawa, A, Kakita, K, Yamada, T, Ishizuka, M, Sakamoto, J, Hatori, N, Maeda, N, Ohsaka, F, Saitoh, T, Nomura, T, Kuroki, K, Nambu, H, Arase, H, Matsunaga, S, Anada, M, Ose, T, Hashimoto, S, Maenaka, K. | | Deposit date: | 2017-05-25 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and thermodynamic analyses reveal critical features of glycopeptide recognition by the human PILR alpha immune cell receptor.
J. Biol. Chem., 292, 2017
|
|
3VU0
 
 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (AfAglB-S2, AF_0040, O30195_ARCFU) from Archaeoglobus fulgidus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative uncharacterized protein | | Authors: | Nyirenda, J, Matsumoto, S, Saitoh, T, Maita, N, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystallographic and NMR Evidence for Flexibility in Oligosaccharyltransferases and Its Catalytic Significance
Structure, 21, 2013
|
|
3VU1
 
 | | Crystal structure of the C-terminal globular domain of oligosaccharyltransferase (PhAglB-L, O74088_PYRHO) from Pyrococcus horikoshii | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative uncharacterized protein PH0242 | | Authors: | Nyirenda, J, Matsumoto, S, Saitoh, T, Maita, N, Noda, N.N, Inagaki, F, Kohda, D. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic and NMR Evidence for Flexibility in Oligosaccharyltransferases and Its Catalytic Significance
Structure, 21, 2013
|
|
2V1S
 
 | | CRYSTAL STRUCTURE OF RAT TOM20-ALDH PRESEQUENCE COMPLEX | | Descriptor: | ALDEHYDE DEHYDROGENASE, MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20 HOMOLOG | | Authors: | Obita, T, Igura, M, Ose, T, Endo, T, Maenaka, K, Kohda, D. | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tom20 Recognizes Mitochondrial Presequences Through Dynamic Equilibrium Among Multiple Bound States.
Embo J., 26, 2007
|
|
2V1T
 
 | | CRYSTAL STRUCTURE OF RAT TOM20-ALDH PRESEQUENCE COMPLEX | | Descriptor: | ALDEHYDE DEHYDROGENASE, MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20 HOMOLOG | | Authors: | Obita, T, Igura, M, Ose, T, Endo, T, Maenaka, K, Kohda, D. | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Tom20 Recognizes Mitochondrial Presequences Through Dynamic Equilibrium Among Multiple Bound States.
Embo J., 26, 2007
|
|
3KGL
 
 | | Crystal structure of procruciferin, 11S globulin from Brassica napus | | Descriptor: | Cruciferin, GLYCEROL, SULFATE ION | | Authors: | Tandang-Silvas, M.R, Mikami, B, Maruyama, N, Utsumi, S. | | Deposit date: | 2009-10-29 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 1804, 2010
|
|
6LX7
 
 | | X-ray structure of human PPARalpha ligand binding domain-stearic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, STEARIC ACID | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LXC
 
 | | X-ray structure of human PPARalpha ligand binding domain-saroglitazar co-crystals obtained by delipidation and cross-seeding | | Descriptor: | (2S)-2-ethoxy-3-[4-[2-[2-methyl-5-(4-methylsulfanylphenyl)pyrrol-1-yl]ethoxy]phenyl]propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Honda, A, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L37
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-ciprofibrate co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-chloro-5-nitro-N-phenylbenzamide, 2-{4-[(1S)-2,2-dichlorocyclopropyl]phenoxy}-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L36
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
