4YCR
 
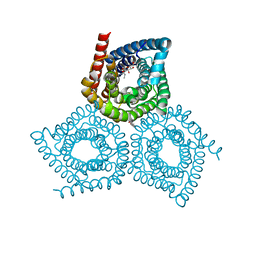 | | Structure determination of an integral membrane protein at room temperature from crystals in situ | | Descriptor: | Tellurite resistance protein TehA homolog, octyl beta-D-glucopyranoside | | Authors: | Axford, D, Hu, N.J, Foadi, J, Choudhury, H.G, Iwata, S, Beis, K, Evans, G, Alguel, Y. | | Deposit date: | 2015-02-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure determination of an integral membrane protein at room temperature from crystals in situ.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1XIP
 
 | |
1Q51
 
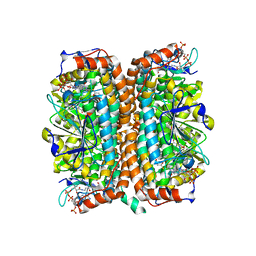 | | Crystal Structure of Mycobacterium tuberculosis MenB in Complex with Acetoacetyl-Coenzyme A, a Key Enzyme in Vitamin K2 Biosynthesis | | Descriptor: | ACETOACETYL-COENZYME A, menB | | Authors: | Truglio, J.J, Theis, K, Feng, Y, Gajda, R, Machutta, C, Tonge, P.J, Kisker, C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-05 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
J.Biol.Chem., 278, 2003
|
|
1Q52
 
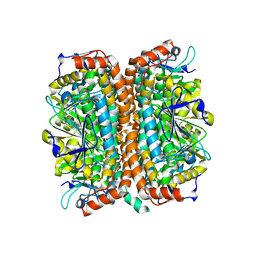 | | Crystal Structure of Mycobacterium tuberculosis MenB, a Key Enzyme in Vitamin K2 Biosynthesis | | Descriptor: | menB | | Authors: | Truglio, J.J, Theis, K, Feng, Y, Gajda, R, Machutta, C, Tonge, P.J, Kisker, C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-05 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
J.Biol.Chem., 278, 2003
|
|
2KY7
 
 | | NMR Structural Studies on the Covalent DNA Binding of a Pyrrolobenzodiazepine-Naphthalimide Conjugate | | Descriptor: | 2-{2-[4-(3-{[(11aS)-7-methoxy-5-oxo-2,3,5,10,11,11a-hexahydro-1H-pyrrolo[2,1-c][1,4]benzodiazepin-8-yl]oxy}propyl)piperazin-1-yl]ethyl}-1H-benzo[de]isoquinoline-1,3(2H)-dione, 5'-D(*AP*AP*CP*AP*AP*TP*TP*GP*TP*T)-3' | | Authors: | Rettig, M, Langel, W, Kamal, A, Weisz, K. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies on the covalent DNA binding of a pyrrolobenzodiazepine-naphthalimide conjugate
Org.Biomol.Chem., 8, 2010
|
|
1T5L
 
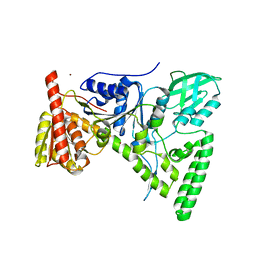 | | Crystal structure of the DNA repair protein UvrB point mutant Y96A revealing a novel fold for domain 2 | | Descriptor: | UvrABC system protein B, ZINC ION | | Authors: | Truglio, J.J, Croteau, D.L, Skorvaga, M, DellaVecchia, M.J, Theis, K, Mandavilli, B.S, Van Houten, B, Kisker, C. | | Deposit date: | 2004-05-04 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactions between UvrA and UvrB: the role of UvrB's domain 2 in nucleotide excision repair
Embo J., 23, 2004
|
|
1JRO
 
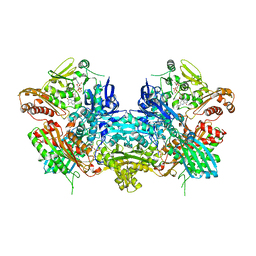 | | Crystal Structure of Xanthine Dehydrogenase from Rhodobacter capsulatus | | Descriptor: | CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Truglio, J.J, Theis, K, Leimkuhler, S, Rappa, R, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2001-08-14 | | Release date: | 2002-01-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the active and alloxanthine-inhibited forms of xanthine dehydrogenase from Rhodobacter capsulatus
Structure, 10, 2002
|
|
1JRP
 
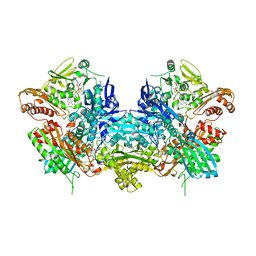 | | Crystal Structure of Xanthine Dehydrogenase inhibited by alloxanthine from Rhodobacter capsulatus | | Descriptor: | CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Truglio, J.J, Theis, K, Leimkuhler, S, Rappa, R, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2001-08-14 | | Release date: | 2002-01-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the active and alloxanthine-inhibited forms of xanthine dehydrogenase from Rhodobacter capsulatus
Structure, 10, 2002
|
|
1KET
 
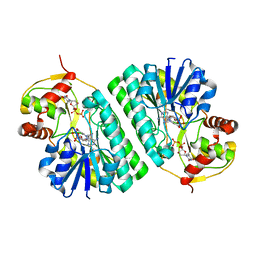 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with thymidine diphosphate bound | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, THYMIDINE-5'-DIPHOSPHATE, dTDP-D-glucose 4,6-dehydratase | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
1KEU
 
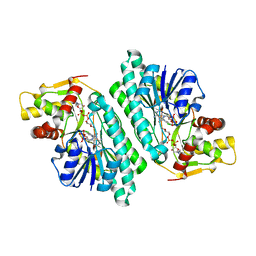 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium with dTDP-D-glucose bound | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, dTDP-D-glucose 4,6-dehydratase | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
1KEP
 
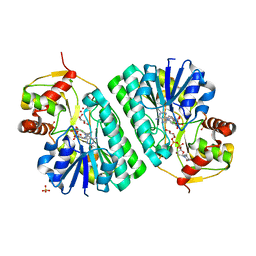 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with dTDP-xylose bound | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, THYMIDINE-5'-DIPHOSPHO-BETA-D-XYLOSE, ... | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-16 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
1KEW
 
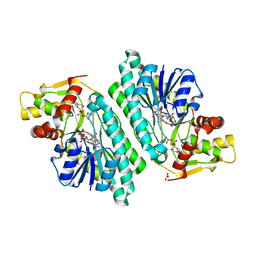 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium with thymidine diphosphate bound | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
1KER
 
 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with dTDP-D-glucose bound | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
1G5H
 
 | | CRYSTAL STRUCTURE OF THE ACCESSORY SUBUNIT OF MURINE MITOCHONDRIAL POLYMERASE GAMMA | | Descriptor: | GLYCEROL, MITOCHONDRIAL DNA POLYMERASE ACCESSORY SUBUNIT, SODIUM ION | | Authors: | Carrodeguas, J.A, Theis, K, Bogenhagen, D.F, Kisker, C. | | Deposit date: | 2000-11-01 | | Release date: | 2001-03-14 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer.
Mol.Cell, 7, 2001
|
|
1G5I
 
 | | CRYSTAL STRUCTURE OF THE ACCESSORY SUBUNIT OF MURINE MITOCHONDRIAL POLYMERASE GAMMA | | Descriptor: | GLYCEROL, MITOCHONDRIAL DNA POLYMERASE ACCESSORY SUBUNIT, SODIUM ION | | Authors: | Carrodeguas, J.A, Theis, K, Bogenhagen, D.F, Kisker, C. | | Deposit date: | 2000-11-01 | | Release date: | 2001-03-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer.
Mol.Cell, 7, 2001
|
|
2XVA
 
 | | Crystal structure of the tellurite detoxification protein TehB from E. coli in complex with sinefungin | | Descriptor: | SINEFUNGIN, TELLURITE RESISTANCE PROTEIN TEHB, ZINC ION | | Authors: | Choudhury, H.G, Cameron, A.D, Iwata, S, Beis, K. | | Deposit date: | 2010-10-25 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Mechanism of the Chalcogen Detoxifying Protein Tehb from Escherichia Coli.
Biochem.J., 435, 2011
|
|
2XVM
 
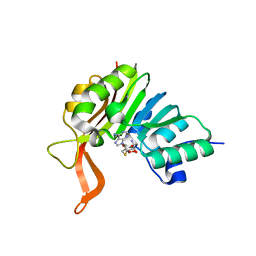 | | Crystal structure of the tellurite detoxification protein TehB from E. coli in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, TELLURITE RESISTANCE PROTEIN TEHB | | Authors: | Choudhury, H.G, Cameron, A.D, Iwata, S, Beis, K. | | Deposit date: | 2010-10-26 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure and Mechanism of the Chalcogen Detoxifying Protein Tehb from Escherichia Coli.
Biochem.J., 435, 2011
|
|
5MBR
 
 | |
5MCR
 
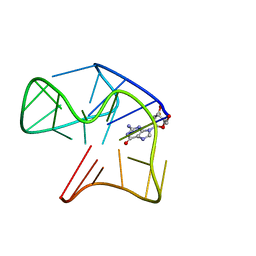 | | Quadruplex with flipped tetrad formed by an artificial sequence | | Descriptor: | Artificial quadruplex with propeller, diagonal and lateral loop | | Authors: | Dickerhoff, J, Haase, L, Langel, W, Weisz, K. | | Deposit date: | 2016-11-10 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Tracing Effects of Fluorine Substitutions on G-Quadruplex Conformational Changes.
ACS Chem. Biol., 12, 2017
|
|
5OV2
 
 | |
4ADO
 
 | | Fusidic acid resistance protein FusB | | Descriptor: | FAR1, ZINC ION | | Authors: | Guo, X, Peisker, K, Backbro, K, Chen, Y, Kiran, R.K, Sanyal, S, Selmer, M. | | Deposit date: | 2011-12-31 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of Fusb: An Elongation Factor G-Binding Fusidic Acid Resistance Protein Active in Ribosomal Translocation and Recycling
Open Biol., 2, 2012
|
|
4ADN
 
 | | Fusidic acid resistance protein FusB | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FAR1, ... | | Authors: | Guo, X, Peisker, K, Backbro, K, Chen, Y, Kiran, R.K, Sanyal, S, Selmer, M. | | Deposit date: | 2011-12-31 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Function of Fusb: An Elongation Factor G-Binding Fusidic Acid Resistance Protein Active in Ribosomal Translocation and Recycling
Open Biol., 2, 2012
|
|
4CU4
 
 | | FhuA from E. coli in complex with the lasso peptide microcin J25 (MccJ25) | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 3-HYDROXY-TETRADECANOIC ACID, DIPHOSPHATE, ... | | Authors: | Mathavan, I, Rebuffat, S, Beis, K. | | Deposit date: | 2014-03-17 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Hijacking Siderophore Receptors by Antimicrobial Lasso Peptides.
Nat.Chem.Biol., 10, 2014
|
|
4B09
 
 | | Structure of unphosphorylated BaeR dimer | | Descriptor: | HEXATANTALUM DODECABROMIDE, TRANSCRIPTIONAL REGULATORY PROTEIN BAER | | Authors: | Choudhury, H, Beis, K. | | Deposit date: | 2012-06-29 | | Release date: | 2013-07-10 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Dimeric Form of the Unphosphorylated Response Regulator Baer.
Protein Sci., 22, 2013
|
|
5IWL
 
 | | CD47-diabody complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5F9 diabody, Leukocyte surface antigen CD47, ... | | Authors: | Di, W, Jude, K.M, Garcia, K.C. | | Deposit date: | 2016-03-22 | | Release date: | 2016-06-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer.
J.Clin.Invest., 126, 2016
|
|
