4OUM
 
 | | Crystal structure of human Caprin-2 C1q domain | | Descriptor: | CITRATE ANION, Caprin-2, ISOPROPYL ALCOHOL, ... | | Authors: | Song, X, Li, L. | | Deposit date: | 2014-02-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
J.Biol.Chem., 289, 2014
|
|
4OUL
 
 | | Crystal structure of human Caprin-2 C1q domain | | Descriptor: | CALCIUM ION, Caprin-2, GLYCEROL | | Authors: | Song, X, Li, L. | | Deposit date: | 2014-02-17 | | Release date: | 2014-10-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
J.Biol.Chem., 289, 2014
|
|
4OUS
 
 | | Crystal structure of zebrafish Caprin-2 C1q domain | | Descriptor: | CALCIUM ION, Caprin-2 | | Authors: | Song, X, Li, L. | | Deposit date: | 2014-02-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
J.Biol.Chem., 289, 2014
|
|
6GEL
 
 | | The structure of TWITCH-2B | | Descriptor: | CALCIUM ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-26 | | Release date: | 2019-08-21 | | Last modified: | 2019-09-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
6GEZ
 
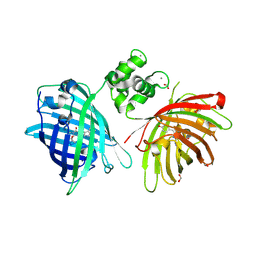 | | THE STRUCTURE OF TWITCH-2B N532F | | Descriptor: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Optimized Ratiometric Calcium Sensor,Green fluorescent protein,Green fluorescent protein | | Authors: | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | Deposit date: | 2018-04-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
2M97
 
 | |
6O3X
 
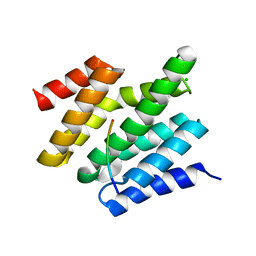 | | Crystal structure of yeast Nrd1 CID in complex with Sen1 NIM2 | | Descriptor: | CHLORIDE ION, Helicase SEN1, Protein NRD1 | | Authors: | Zhang, Y, Tong, L. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Identification of Three Sequence Motifs in the Transcription Termination Factor Sen1 that Mediate Direct Interactions with Nrd1.
Structure, 27, 2019
|
|
6O3W
 
 | |
6O3Y
 
 | | Crystal structure of yeast Nrd1 CID in complex with Sen1 NIM3 | | Descriptor: | CHLORIDE ION, Helicase SEN1, Protein NRD1 | | Authors: | Zhang, Y, Tong, L. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Identification of Three Sequence Motifs in the Transcription Termination Factor Sen1 that Mediate Direct Interactions with Nrd1.
Structure, 27, 2019
|
|
2D6F
 
 | |
6NNW
 
 | | Tsn15 in complex with substrate intermediate | | Descriptor: | (3E)-3-{(1S,4S,4aS,5R,8aS)-1-[(2E,4R,7S,8E,10S)-1,7-dihydroxy-10-{(2R,3S,5R)-5-[(1S)-1-methoxyethyl]-3-methyloxolan-2-yl}-4-methylundeca-2,8-dien-2-yl]-4,5-dimethyloctahydro-3H-2-benzopyran-3-ylidene}oxolane-2,4-dione, PHOSPHATE ION, PROPANOIC ACID, ... | | Authors: | Paiva, F.C.R, Little, R, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2019-01-15 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unexpected enzyme-catalysed [4+2] cycloaddition and rearrangement in polyether antibiotic biosynthesis
Nat Catal, 2019
|
|
6NOI
 
 | | Crystal structure of Tsn15 in apo form | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Little, R, Paiva, F.C.R, Dias, M.V.B, Leadlay, P. | | Deposit date: | 2019-01-16 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unexpected enzyme-catalysed [4+2] cycloaddition and rearrangement in polyether antibiotic biosynthesis
Nat Catal, 2019
|
|
7QO8
 
 | |
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
1MEA
 
 | |
1MED
 
 | |
3B2R
 
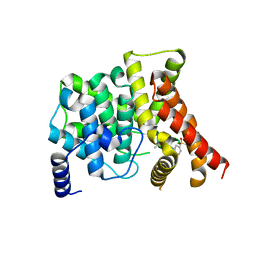 | | Crystal Structure of PDE5A1 catalytic domain in complex with Vardenafil | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Huanchen, W, Mengchun, Y, Howard, R, Sharron, H.F, Hengming, K. | | Deposit date: | 2007-10-19 | | Release date: | 2008-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Conformational variations of both phosphodiesterase-5 and inhibitors provide the structural basis for the physiological effects of vardenafil and sildenafil.
Mol.Pharmacol., 73, 2008
|
|
