4QLC
 
 | | Crystal structure of chromatosome at 3.5 angstrom resolution | | Descriptor: | CITRIC ACID, DNA (167-mer), H5, ... | | Authors: | Jiang, J.S, Zhou, B.R, Xiao, T.S, Bai, Y.W. | | Deposit date: | 2014-06-11 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structural Mechanisms of Nucleosome Recognition by Linker Histones.
Mol.Cell, 33 Suppl 1, 2015
|
|
7K79
 
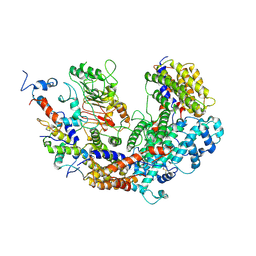 | | CBF3 | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, Suppressor of kinetochore protein 1 | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
7K7G
 
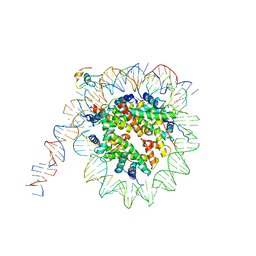 | | nucleosome and Gal4 complex | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, DNA (147-MER), Histone H2A.1, ... | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
7K78
 
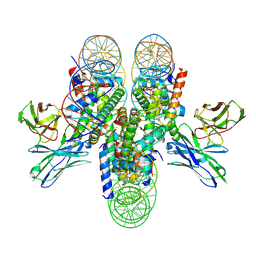 | | antibody and nucleosome complex | | Descriptor: | Cse4, DNA (136-MER), Histone H2A.1, ... | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
7CHF
 
 | |
7CHH
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with BD-368-2 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-368-2 Fab heavy chain, ... | | Authors: | Xiao, J, Zhu, Q, Wang, G. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
8HKG
 
 | |
7CH5
 
 | |
7CHE
 
 | |
7CH4
 
 | |
7CHB
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with BD-236 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-236 Fab heavy chain, BD-236 Fab light chain, ... | | Authors: | Xiao, J, Zhu, Q. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
7CHC
 
 | |
7EXA
 
 | | Structure of mumps virus nucleoprotein without C-arm | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Shen, Q, Shan, H, Zhang, N, Qin, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural plasticity of mumps virus nucleocapsids with cryo-EM structures.
Commun Biol, 4, 2021
|
|
7EWQ
 
 | | Structure of Mumps virus nucleocapsid ring | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Su, X, Shen, Q, Shan, H. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-23 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural plasticity of mumps virus nucleocapsids with cryo-EM structures.
Commun Biol, 4, 2021
|
|
4R0T
 
 | | Crystal structure of P. aeruginosa TpbA (C132S) in complex with pTyr | | Descriptor: | PHOSPHATE ION, Protein tyrosine phosphatase TpbA, TYROSINE | | Authors: | Xu, K, Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1
Plos One, 10
|
|
6CYT
 
 | | HIV-1 TAR loop in complex with Tat:AFF4:P-TEFb | | Descriptor: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Schulze Gahmen, U, Hurley, J.H. | | Deposit date: | 2018-04-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural mechanism for HIV-1 TAR loop recognition by Tat and the super elongation complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6V0N
 
 | | PRMT5 bound to PBM peptide from Riok1 | | Descriptor: | Methylosome protein 50, Protein arginine N-methyltransferase 5, Riok1 PBM peptide, ... | | Authors: | McMIllan, B.J, Raymond, D.D. | | Deposit date: | 2019-11-19 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Molecular basis for substrate recruitment to the PRMT5 methylosome.
Mol.Cell, 81, 2021
|
|
6V0O
 
 | | PRMT5 bound to the PBM peptide from pICln | | Descriptor: | ACETYL GROUP, Methylosome protein 50, PBM peptide, ... | | Authors: | McMillan, B.J, Raymond, D.D. | | Deposit date: | 2019-11-19 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Molecular basis for substrate recruitment to the PRMT5 methylosome.
Mol.Cell, 81, 2021
|
|
