5XWS
 
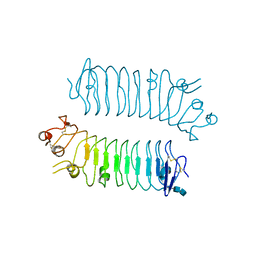 | | Crystal structure of SALM5 LRR-Ig | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich repeat and fibronectin type-III domain-containing protein 5 | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.084 Å) | | Cite: | Structural basis of trans-synaptic interactions between PTP delta and SALMs for inducing synapse formation.
Nat Commun, 9, 2018
|
|
5XWT
 
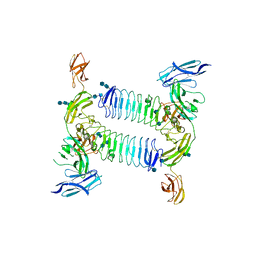 | | Crystal structure of PTPdelta Ig1-Fn1 in complex with SALM5 LRR-Ig | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.178 Å) | | Cite: | Structural basis of trans-synaptic interactions between PTP delta and SALMs for inducing synapse formation.
Nat Commun, 9, 2018
|
|
3KBJ
 
 | |
1GCU
 
 | |
3KCJ
 
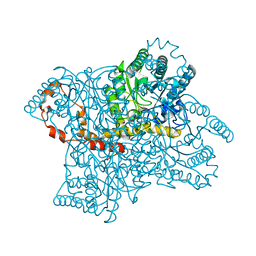 | |
5Y32
 
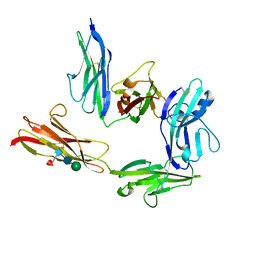 | | Crystal structure of PTP delta Ig1-Ig2 in complex with IL1RAPL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2017-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Mechanisms of splicing-dependent trans-synaptic adhesion by PTP delta-IL1RAPL1/IL-1RAcP for synaptic differentiation.
Nat Commun, 6, 2015
|
|
1IW0
 
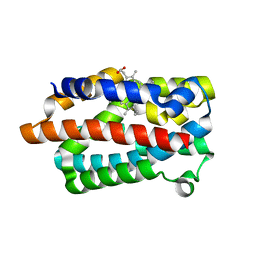 | | Crystal structure of a heme oxygenase (HmuO) from Corynebacterium diphtheriae complexed with heme in the ferric state | | Descriptor: | Heme oxygenase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Hirotsu, S, Unno, M, Chu, G.C, Lee, D.S, Park, S.Y, Shiro, Y, Ikeda-Saito, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-04-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of the ferric and ferrous forms of the heme complex of HmuO, a heme oxygenase of Corynebacterium diphtheriae.
J.Biol.Chem., 279, 2004
|
|
5B1H
 
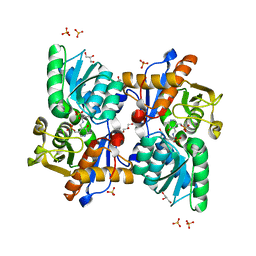 | |
5B1I
 
 | |
2E5B
 
 | |
2E5D
 
 | | Crystal structure of Human NMPRTase complexed with nicotinamide | | Descriptor: | NICOTINAMIDE, Nicotinamide phosphoribosyltransferase | | Authors: | Takahashi, R, Nakamura, S, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase
J.Biochem., 147, 2010
|
|
2E5C
 
 | | Crystal structure of Human NMPRTase complexed with 5'-phosphoribosyl-1'-pyrophosphate | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Nicotinamide phosphoribosyltransferase | | Authors: | Takahashi, R, Nakamura, S, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase
J.Biochem., 147, 2010
|
|
2DKN
 
 | |
