9AU2
 
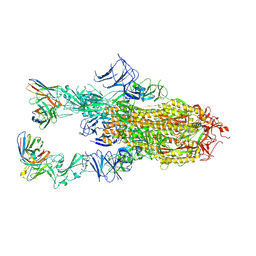 | | VIR-7229 Fab fragment bound the BA.2.86 spike trimer (global refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Park, Y.J, Veelser, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
8F82
 
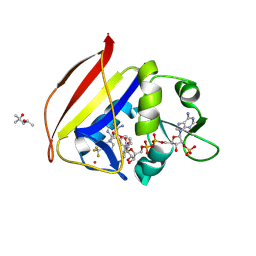 | | Crystal Structure of Dihydrofolate reductase (DHFR) from Mycobacterium ulcerans Agy99 in complex with NADP and inhibitor MAM845 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-{2-[3-({(6M)-2,4-diamino-6-[3-(trifluoromethyl)phenyl]pyrimidin-5-yl}oxy)propoxy]phenyl}propanoic acid, BROMIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-21 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rational Exploration of 2,4-Diaminopyrimidines as DHFR Inhibitors Active against Mycobacterium abscessus and Mycobacterium avium , Two Emerging Human Pathogens.
J.Med.Chem., 67, 2024
|
|
8F83
 
 | |
8F84
 
 | |
8F80
 
 | | Crystal Structure of Dihydrofolate reductase (DHFR) from Mycobacterium ulcerans Agy99 in complex with NADP and inhibitor MAM738 | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-(3-{[2,4-diamino-6-(4-methoxyphenyl)pyrimidin-5-yl]oxy}propoxy)phenyl]propanoic acid, Dihydrofolate reductase, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-21 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rational Exploration of 2,4-Diaminopyrimidines as DHFR Inhibitors Active against Mycobacterium abscessus and Mycobacterium avium , Two Emerging Human Pathogens.
J.Med.Chem., 67, 2024
|
|
8F85
 
 | |
8F81
 
 | |
8G0V
 
 | |
8G0R
 
 | |
8G0U
 
 | |
8G0T
 
 | |
9AU1
 
 | | SARS-CoV-2 XBB.1.5 RBD bound to the VIR-7229 and the S309 Fab fragments | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Rietz, T, Park, Y.J, Errico, J, Czudnochowski, N, Nix, J.C, Corti, D, Snell, G, Marco, A.D, Pinto, D, Cameroni, E, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-02-27 | | Release date: | 2024-10-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 187, 2024
|
|
4YLG
 
 | |
8VJ2
 
 | |
5E3I
 
 | |
6MJN
 
 | |
4OL9
 
 | |
8DQC
 
 | |
8DQ9
 
 | |
8DQB
 
 | |
8DU0
 
 | |
7RA8
 
 | |
7RAL
 
 | |
8F8U
 
 | |
8FI4
 
 | |
