6U6C
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate- and GSK2-bound form | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-fluorobenzene-1-carbonyl)-N-methyl-2,3-dihydro-1H-indole-5-sulfonamide, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Allosteric inhibitors of Mycobacterium tuberculosis tryptophan synthase.
Protein Sci., 29, 2020
|
|
6U0Y
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia | | Descriptor: | 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia
To Be Published
|
|
4ICH
 
 | | Crystal structure of a putative TetR family transcriptional regulator from Saccharomonospora viridis DSM 43017 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-02 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a putative TetR family transcriptional regulator from Saccharomonospora viridis DSM 43017
To be Published
|
|
6UA1
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the no-metal bound form | | Descriptor: | 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase) | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the no-metal bound form.
To Be Published
|
|
6UAF
 
 | | Crystal Structure of the Metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Imipnem | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Imipnem
To Be Published
|
|
6U9C
 
 | | The 2.2 A Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the complex with Acetyl CoA | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-07 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the complex with Acetyl CoA
To Be Published
|
|
6UB9
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - AMINOACRYLATE- AND BRD6309-BOUND FORM | | Descriptor: | (2R,3S,4R)-3-(4'-chloro-2',6'-difluoro[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-11 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - AMINOACRYLATE- AND BRD6309-BOUND FORM
To be Published
|
|
6UHH
 
 | | Crystal Structure of Human RYR Receptor 3 ( 848-1055) in Complex with ATP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, R, Kim, Y, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-09-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.138 Å) | | Cite: | Crystal Structure of Human RYR Receptor 3 ( 848-1055) in Complex with ATP
To Be Published
|
|
4IPT
 
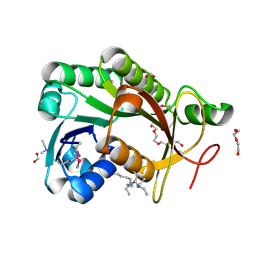 | | The crystal structure of a short-chain dehydrogenases/reductase (ethylated) from Veillonella parvula DSM 2008 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-10 | | Release date: | 2013-02-06 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | The crystal structure of a short-chain dehydrogenases/reductase (ethylated) from Veillonella parvula DSM 2008
To be Published
|
|
4JGP
 
 | | The crystal structure of sporulation kinase D sensor domain from Bacillus subtilis subsp in complex with pyruvate at 2.0A resolution | | Descriptor: | PYRUVIC ACID, Sporulation kinase D | | Authors: | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
6UHA
 
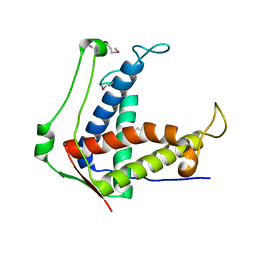 | |
4JNN
 
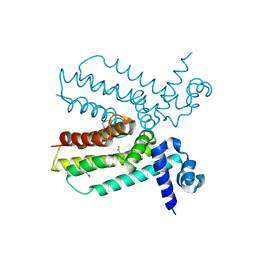 | | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with benzamidine | | Descriptor: | BENZAMIDINE, BETA-MERCAPTOETHANOL, Transcriptional regulator | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-15 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with benzamidine
To be Published
|
|
6UHB
 
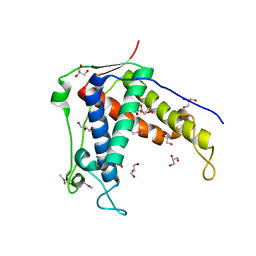 | |
6UAH
 
 | | Crystal Structure of the Metallo-beta-Lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of the Metallo-beta-Lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Meropenem
To Be Published
|
|
6UAM
 
 | |
4KWA
 
 | | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with choline | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, CHOLINE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with choline
To be Published
|
|
6UHE
 
 | |
6UG5
 
 | |
4JWO
 
 | | The crystal structure of a possible phosphate binding protein from Planctomyces limnophilus DSM 3776 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Tan, K, Gu, M, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-27 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The crystal structure of a possible phosphate binding protein from Planctomyces limnophilus DSM 3776
To be Published
|
|
6UHI
 
 | |
6USA
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate- and GSK1-bound form | | Descriptor: | (3R,4R)-4-[4-(2-Chlorophenyl)piperazin-1-yl]-1,1-dioxothiolan-3-ol, 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-10-25 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Allosteric inhibitors of Mycobacterium tuberculosis tryptophan synthase.
Protein Sci., 29, 2020
|
|
6V70
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Cadmium in the Active Site | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CADMIUM ION, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica.
To Be Published
|
|
6UR7
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-10-22 | | Release date: | 2020-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.709 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
4KVF
 
 | | The crystal structure of a rhamnose ABC transporter, periplasmic rhamnose-binding protein from Kribbella flavida DSM 17836 | | Descriptor: | GLYCEROL, Rhamnose ABC transporter, periplasmic rhamnose-binding protein | | Authors: | Tan, K, Hatzos-Skintges, C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-22 | | Release date: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | The crystal structure of a rhamnose ABC transporter, periplasmic rhamnose-binding protein from Kribbella flavida DSM 17836
To be Published
|
|
4JBF
 
 | | Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469. | | Descriptor: | Peptidoglycan glycosyltransferase, TETRAETHYLENE GLYCOL | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469.
To be Published
|
|
