4DDF
 
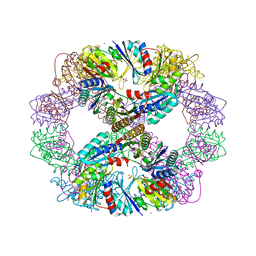 | | Computationally Designed Self-assembling Octahedral Cage protein, O333, Crystallized in space group P4 | | Descriptor: | CHLORIDE ION, Propanediol utilization polyhedral body protein PduT, SULFATE ION | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
4DHX
 
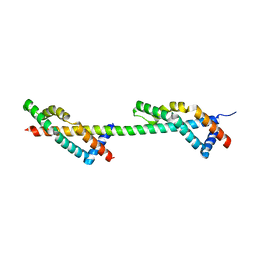 | | ENY2:GANP complex | | Descriptor: | 80 kDa MCM3-associated protein, Enhancer of yellow 2 transcription factor homolog | | Authors: | Stewart, M, Jani, D. | | Deposit date: | 2012-01-30 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export.
Nucleic Acids Res., 40, 2012
|
|
6F77
 
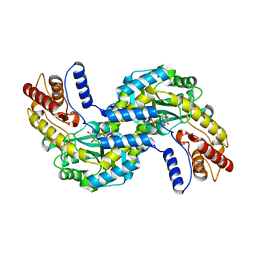 | | Crystal structure of the prephenate aminotransferase from Rhizobium meliloti | | Descriptor: | Aspartate aminotransferase A, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cobessi, D, Giustini, C, Graindorge, M, Matringe, M. | | Deposit date: | 2017-12-07 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Febs J., 286, 2019
|
|
6EYA
 
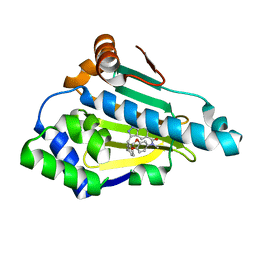 | |
6FEA
 
 | | A. vinelandii vanadium nitrogenase, turnover state | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CARBONATE ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Sippel, D, Einsle, O. | | Deposit date: | 2017-12-31 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A bound reaction intermediate sheds light on the mechanism of nitrogenase.
Science, 359, 2018
|
|
6FM1
 
 | | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATPanddGMP at 2.3 Angstrom resolution | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2018-01-29 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
6FAP
 
 | | Crystal structure of human BAZ2A PHD zinc finger in complex with Fr23 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2017-12-16 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6FCJ
 
 | |
6EYB
 
 | |
6FGT
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methylpyridinone compound 3 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, ~{N}-[(1-azanylcyclohexyl)methyl]-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6FHP
 
 | | DAIP in complex with a C-terminal fragment of thermolysin | | Descriptor: | Dispase autolysis-inducing protein, Thermolysin | | Authors: | Schmelz, S, Fiebig, D, Fuchsbauer, H.L, Blankenfeldt, W, Scrima, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Destructive twisting of neutral metalloproteases: the catalysis mechanism of the Dispase autolysis-inducing protein from Streptomyces mobaraensis DSM 40487.
FEBS J., 285, 2018
|
|
6EZO
 
 | | Eukaryotic initiation factor EIF2B in complex with ISRIB | | Descriptor: | 2-(4-chloranylphenoxy)-~{N}-[4-[2-(4-chloranylphenoxy)ethanoylamino]cyclohexyl]ethanamide, Human eukaryotic initiation factor EIF2B epsilon subunits, Translation initiation factor eIF-2B subunit alpha, ... | | Authors: | Faille, A, Weis, F, Zyryanova, A, Warren, A.J, Ron, D. | | Deposit date: | 2017-11-16 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Binding of ISRIB reveals a regulatory site in the nucleotide exchange factor eIF2B.
Science, 359, 2018
|
|
6F8E
 
 | | PH domain from TgAPH | | Descriptor: | Pleckstrin homology domain | | Authors: | Darvill, N, Liu, B, Matthews, S, Soldati-Favre, D, Rouse, S, Benjamin, S, Blake, T, Dubois, D.J, Hammoudi, P.M, Pino, P. | | Deposit date: | 2017-12-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Phosphatidic Acid Sensing by APH in Apicomplexan Parasites.
Structure, 26, 2018
|
|
6F1N
 
 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | Descriptor: | 4-[5-[2-aminocarbonyl-3,6-bis(azanyl)-5-cyano-thieno[2,3-b]pyridin-4-yl]-2-methoxy-phenoxy]butanoic acid, Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | Deposit date: | 2017-11-22 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
8S9G
 
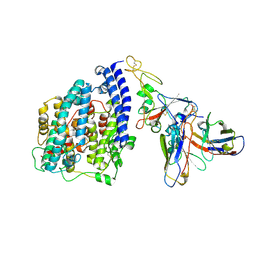 | | SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Fab Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
5HLP
 
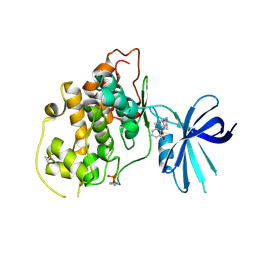 | | X-RAY CRYSTAL STRUCTURE OF GSK3B IN COMPLEX WITH BRD3937 | | Descriptor: | 4-(2-methoxyphenyl)-3,7,7-trimethyl-1,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | White, A, Lakshminarasimhan, D, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inhibitors of Glycogen Synthase Kinase 3 with Exquisite Kinome-Wide Selectivity and Their Functional Effects.
Acs Chem.Biol., 11, 2016
|
|
8VEA
 
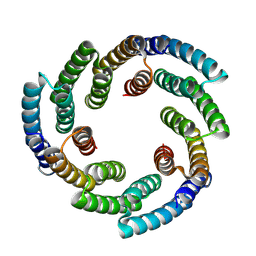 | | De novo designed helical oligomer sg266 | | Descriptor: | Designed helical oligomer sg266 | | Authors: | Bera, A.K, Gerben, S, Baker, D. | | Deposit date: | 2023-12-18 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Massively parallel assessment of designed protein solution properties using mass spectrometry and peptide barcoding.
Biorxiv, 2025
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
8VYE
 
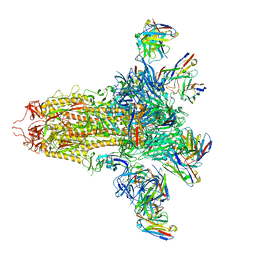 | | SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 Heavy Chain, S2L20 Light Chain, ... | | Authors: | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-02-08 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
8VYF
 
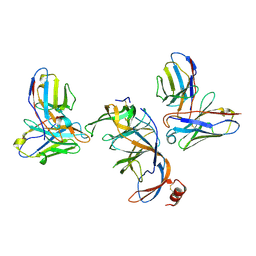 | | SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 Heavy Chain, ... | | Authors: | McCallum, M, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-02-08 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Quantifying how single dose Ad26.COV2.S vaccine efficacy depends on Spike sequence features.
Nat Commun, 15, 2024
|
|
8W22
 
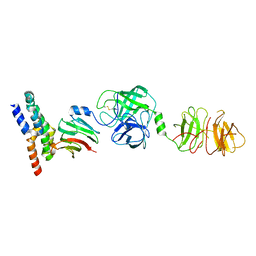 | | Umb1 umbrella toxin particle (local refinement of UmbB1 bound ALF of UmbC1 and UmbA1) | | Descriptor: | Intein C-terminal splicing domain-containing protein, Secreted esterase, Secreted protein | | Authors: | Park, Y.J, Zhao, Q, Seattle Structural Genomics Center for Infectious Disease (SSGCID), DiMaio, F, Mougous, J.D, Veesler, D. | | Deposit date: | 2024-02-19 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Streptomyces umbrella toxin particles block hyphal growth of competing species.
Nature, 629, 2024
|
|
8W20
 
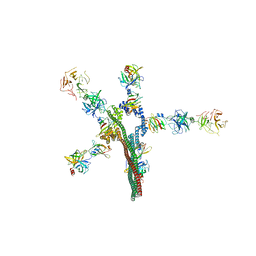 | | Umb1 umbrella toxin particle | | Descriptor: | Intein C-terminal splicing domain-containing protein, Secreted esterase, Secreted protein | | Authors: | Park, Y.J, Zhao, Q, Seattle Structural Genomics Center for Infectious Disease (SSGCID), DiMaio, F, Mougous, J.D, Veesler, D. | | Deposit date: | 2024-02-19 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Streptomyces umbrella toxin particles block hyphal growth of competing species.
Nature, 629, 2024
|
|
8W0S
 
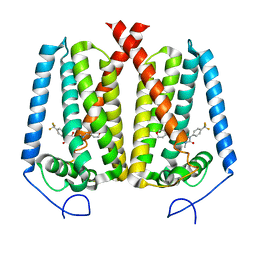 | | Human EBP complexed with compound 3a | | Descriptor: | 1-methyl-8-[(oxan-4-yl)methyl]-3-[4-(trifluoromethyl)phenyl]-1,3,8-triazaspiro[4.5]decane-2,4-dione, 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | | Authors: | Sun, D, Masureel, M. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Discovery and Optimization of Selective Brain-Penetrant EBP Inhibitors that Enhance Oligodendrocyte Formation.
J.Med.Chem., 67, 2024
|
|
8W0R
 
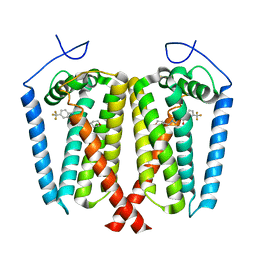 | | Human EBP complexed with compound 1 | | Descriptor: | 1-methyl-1'-[(oxan-4-yl)methyl]-5-(trifluoromethyl)spiro[indole-2,4'-piperidin]-3(1H)-one, 3-beta-hydroxysteroid-Delta(8),Delta(7)-isomerase | | Authors: | Sun, D, Masureel, M. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Discovery and Optimization of Selective Brain-Penetrant EBP Inhibitors that Enhance Oligodendrocyte Formation.
J.Med.Chem., 67, 2024
|
|
8VQ9
 
 | |
