7CSE
 
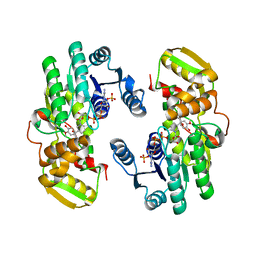 | | AtPrR1 with NADP+ and (-)lariciresinol | | Descriptor: | 4-[[(3S,4S,5R)-4-(hydroxymethyl)-5-(3-methoxy-4-oxidanyl-phenyl)oxolan-3-yl]methyl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.44019055 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS4
 
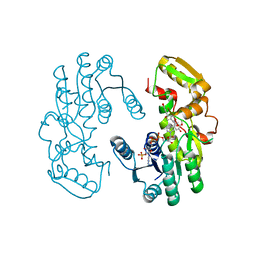 | | IiPLR1 with NADP+ and (+)pinoresinol | | Descriptor: | 4-[(3S,3aR,6S,6aR)-6-(3-methoxy-4-oxidanyl-phenyl)-1,3,3a,4,6,6a-hexahydrofuro[3,4-c]furan-3-yl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.30509257 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS9
 
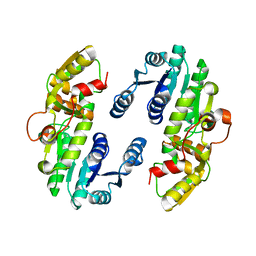 | | AtPrR1 in apo form | | Descriptor: | Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8011415 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSG
 
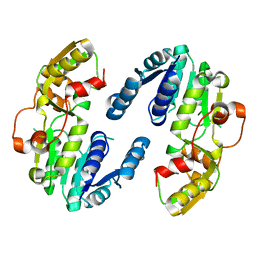 | | AtPrR2 in apo form | | Descriptor: | Pinoresinol reductase 2 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99689388 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSB
 
 | | AtPrR1 with NADP+ and (+)pinoresinol | | Descriptor: | 4-[(3S,3aR,6S,6aR)-6-(3-methoxy-4-oxidanyl-phenyl)-1,3,3a,4,6,6a-hexahydrofuro[3,4-c]furan-3-yl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.002017 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSH
 
 | | AtPrR2 with NADP+ and (+)pinoresinol | | Descriptor: | 4-[(3S,3aR,6S,6aR)-6-(3-methoxy-4-oxidanyl-phenyl)-1,3,3a,4,6,6a-hexahydrofuro[3,4-c]furan-3-yl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 2 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.590775 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS7
 
 | | IiPLR1 with NADP+ and (+)secoisolariciresinol | | Descriptor: | (2S,3S)-2,3-bis[(3-methoxy-4-oxidanyl-phenyl)methyl]butane-1,4-diol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297653 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CSA
 
 | | AtPrR1 with NADP+ | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96212828 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7XP6
 
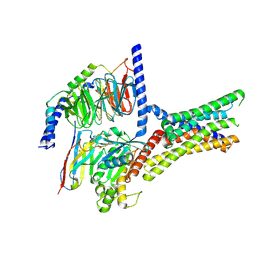 | | Cryo-EM structure of a class T GPCR in active state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP4
 
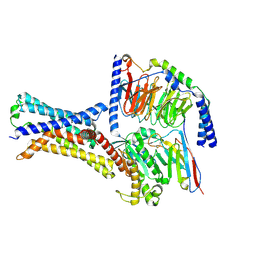 | | Cryo-EM structure of a class T GPCR in apo state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP5
 
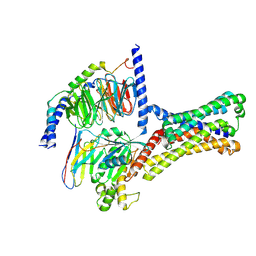 | | Cryo-EM structure of a class T GPCR in ligand-free state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XDI
 
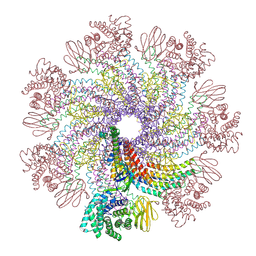 | | Tail structure of bacteriophage SSV19 | | Descriptor: | B210, C131, VP1, ... | | Authors: | Liu, H.R, Chen, W.Y. | | Deposit date: | 2022-03-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into a spindle-shaped archaeal virus with a sevenfold symmetrical tail.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7D7S
 
 | |
7XHF
 
 | |
7XHG
 
 | |
6A82
 
 | |
6AGL
 
 | |
7XXJ
 
 | | Echo 18 incubated with FcRn at pH5.5 | | Descriptor: | PALMITIC ACID, VP1, VP2, ... | | Authors: | Liu, C.C, Qu, X. | | Deposit date: | 2022-05-30 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Human FcRn Is a Two-in-One Attachment-Uncoating Receptor for Echovirus 18.
Mbio, 13, 2022
|
|
7XXA
 
 | | Complex of Echo 18 and FcRn at pH7.4 | | Descriptor: | IgG receptor FcRn large subunit p51, VP1, VP2, ... | | Authors: | Liu, C.C, Qu, X. | | Deposit date: | 2022-05-29 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Human FcRn Is a Two-in-One Attachment-Uncoating Receptor for Echovirus 18.
Mbio, 13, 2022
|
|
7XXG
 
 | | Echo 18 at pH5.5 | | Descriptor: | PALMITIC ACID, VP1, VP2, ... | | Authors: | Liu, C.C, Qu, X. | | Deposit date: | 2022-05-30 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Human FcRn Is a Two-in-One Attachment-Uncoating Receptor for Echovirus 18.
Mbio, 13, 2022
|
|
7Y1R
 
 | | Human L-TGF-beta1 in complex with the anchor protein LRRC33 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duan, Z, Zhang, Z. | | Deposit date: | 2022-06-08 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Specificity of TGF-beta 1 signal designated by LRRC33 and integrin alpha V beta 8.
Nat Commun, 13, 2022
|
|
7Y1T
 
 | | Complex of integrin alphaV/beta8 and L-TGF-beta1 at a ratio of 1:2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duan, Z, Zhang, Z. | | Deposit date: | 2022-06-08 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Specificity of TGF-beta 1 signal designated by LRRC33 and integrin alpha V beta 8.
Nat Commun, 13, 2022
|
|
7DE9
 
 | |
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
6A83
 
 | |
