8K10
 
 | | SID1 transmembrane family member 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K13
 
 | | SID1 transmembrane family member 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K1D
 
 | | SID1 transmembrane family member 1 | | Descriptor: | SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8I5N
 
 | | Rat Kir4.1 in complex with PIP2 and Lys05 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 10, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Zhao, C, Guo, J. | | Deposit date: | 2023-01-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Pharmacological inhibition of Kir4.1 evokes rapid-onset antidepressant responses.
Nat.Chem.Biol., 20, 2024
|
|
7N4X
 
 | | Structure of human NPC1L1 mutant-W347R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-04 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structures of dimeric human NPC1L1 provide insight into mechanisms for cholesterol absorption.
Sci Adv, 7, 2021
|
|
7N4U
 
 | | Structure of human NPC1L1 | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-04 | | Release date: | 2021-09-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structures of dimeric human NPC1L1 provide insight into mechanisms for cholesterol absorption.
Sci Adv, 7, 2021
|
|
6JX1
 
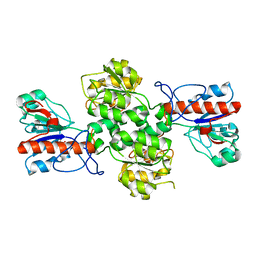 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101 | | Descriptor: | Formate dehydrogenase, GLYCEROL | | Authors: | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | Deposit date: | 2019-04-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
7N8T
 
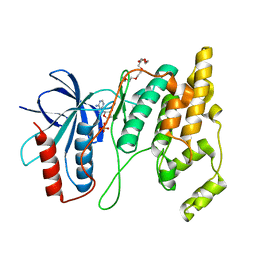 | | Crystal Structure of AMP-bound Human JNK2 | | Descriptor: | ADENOSINE MONOPHOSPHATE, HEXAETHYLENE GLYCOL, Mitogen-activated protein kinase 9 | | Authors: | Li, L, Gurbani, D, Westover, K.D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of a Covalent Inhibitor of c-Jun N-Terminal Protein Kinase (JNK) 2/3 with Selectivity over JNK1.
J.Med.Chem., 66, 2023
|
|
7YC9
 
 | | Co-crystal structure of BTK kinase domain with inhibitor | | Descriptor: | (7~{S})-2-(4-bromanyl-3,5-dimethoxy-phenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, Tyrosine-protein kinase BTK | | Authors: | Zhou, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of BGB-8035, a Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase for B-Cell Malignancies and Autoimmune Diseases.
J.Med.Chem., 66, 2023
|
|
7XO2
 
 | |
7XO0
 
 | |
7XO3
 
 | |
7XO1
 
 | |
4O42
 
 | | Tandem chromodomains of human CHD1 in complex with influenza NS1 C-terminal tail dimethylated at K229 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, GLYCEROL, Nonstructural protein 1, ... | | Authors: | Qin, S, Xu, C, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
6JUK
 
 | | Crystal structure of Formate dehydrogenase mutant C256I/E261P/S381I from Pseudomonas sp. 101 in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | Descriptor: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Feng, Y, Xue, S, Guo, X, Zhao, Z. | | Deposit date: | 2019-04-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JWG
 
 | | Crystal structure of Formate dehydrogenase mutant C256I/E261P/S381I from Pseudomonas sp. 101 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Formate dehydrogenase, GLYCEROL | | Authors: | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | Deposit date: | 2019-04-20 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
6JUJ
 
 | | Crystal structure of Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F from Pseudomonas sp. 101in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | Descriptor: | Formate dehydrogenase, GLYCEROL, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Feng, Y, Guo, X, Xue, S, Zhao, Z. | | Deposit date: | 2019-04-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Structure-Guided Design of Formate Dehydrogenase for Regeneration of a Non-Natural Redox Cofactor.
Chemistry, 26, 2020
|
|
5WRW
 
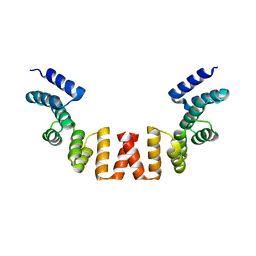 | | Structure of human apo-SRP72 | | Descriptor: | SULFATE ION, Signal recognition particle subunit SRP72 | | Authors: | Gao, Y, Chen, Z. | | Deposit date: | 2016-12-04 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Human apo-SRP72 and SRP68/72 complex structures reveal the molecular basis of protein translocation
J Mol Cell Biol, 9, 2017
|
|
4GJ3
 
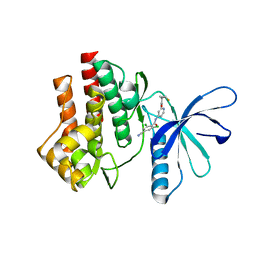 | | Tyk2 (JH1) in complex with 2,6-dichloro-4-cyano-N-[2-({[(1R,2R)-2-fluorocyclopropyl]carbonyl}amino)pyridin-4-yl]benzamide | | Descriptor: | 2,6-dichloro-4-cyano-N-[2-({[(1R,2R)-2-fluorocyclopropyl]carbonyl}amino)pyridin-4-yl]benzamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Ultsch, M.H. | | Deposit date: | 2012-08-09 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lead Optimization of a 4-Aminopyridine Benzamide Scaffold To Identify Potent, Selective, and Orally Bioavailable TYK2 Inhibitors.
J.Med.Chem., 56, 2013
|
|
5XMZ
 
 | | Verticillium effector PevD1 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Effector protein PevD1 | | Authors: | Liu, X, Zhou, R. | | Deposit date: | 2017-05-17 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The asparagine-rich protein NRP interacts with the Verticillium effector PevD1 and regulates the subcellular localization of cryptochrome 2
J. Exp. Bot., 68, 2017
|
|
8JCC
 
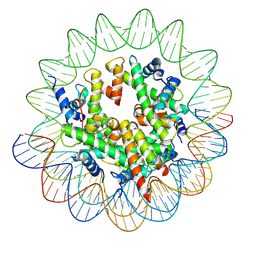 | |
8JCD
 
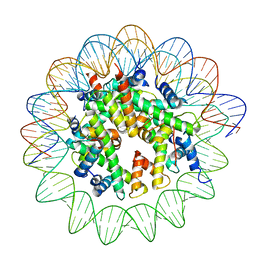 | | Human H2BFWTH100R nucleosome with 601 DNA | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type W-T, ... | | Authors: | Ding, D.B, Ishibashi, T, Nguyen, T.T. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Testis-specific H2B.W1 disrupts nucleosome integrity by reducing DNA-histone interactions.
Nucleic Acids Res., 2024
|
|
8JBX
 
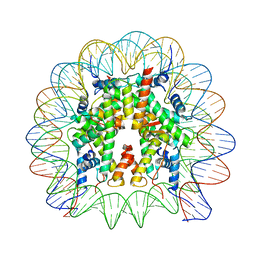 | | Human canonical 601 DNA nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Ding, D.B, Ishibashi, T, Nguyen, T.T. | | Deposit date: | 2023-05-09 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Testis-specific H2B.W1 disrupts nucleosome integrity by reducing DNA-histone interactions.
Nucleic Acids Res., 2024
|
|
5CJB
 
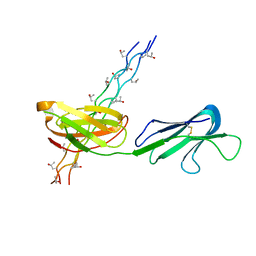 | | Human Osteoclast Associated Receptor (OSCAR) in complex with a collagen-like peptide | | Descriptor: | Osteoclast-associated immunoglobulin-like receptor, collagen-like peptide | | Authors: | Gao, G.F, Qi, J, Shi, Y, Haywood, J. | | Deposit date: | 2015-07-14 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis of collagen recognition by human osteoclast-associated receptor and design of osteoclastogenesis inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8GUN
 
 | |
