4MPY
 
 | | 1.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus (IDP00699) in complex with NAD+ | | Descriptor: | Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-14 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based mutational studies of substrate inhibition of betaine aldehyde dehydrogenase BetB from Staphylococcus aureus.
Appl.Environ.Microbiol., 80, 2014
|
|
4JFR
 
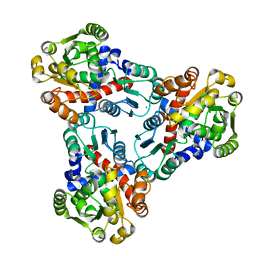 | | Crystal structure of anabolic ornithine carbamoyltransferase from Vibrio vulnificus in complex with carbamoyl phosphate | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Ornithine carbamoyltransferase, ... | | Authors: | Shabalin, I.G, Winsor, J, Grimshaw, S, Osinski, T, Bajor, J, Chordia, M.D, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
to be published
|
|
3EFB
 
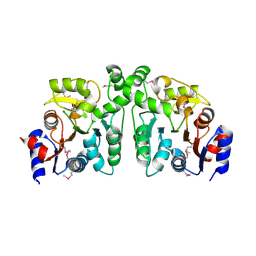 | | Crystal Structure of Probable sor Operon Regulator from Shigella flexneri | | Descriptor: | ACETIC ACID, Probable sor-operon regulator | | Authors: | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Probable sor Operon Regulator from Shigella flexneri
To be Published
|
|
4QM1
 
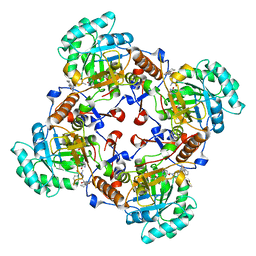 | | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor D67 | | Descriptor: | 2-(3-methyl-4-oxo-3,4-dihydrophthalazin-1-yl)-N-(6,7,8,9-tetrahydrodibenzo[b,d]furan-2-yl)acetamide, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Mandapati, K, Gollapalli, D, Gorla, S.K, Zhang, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-14 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7964 Å) | | Cite: | Crystal Structure of the Inosine 5'-monophosphate Dehydrogenase with an Internal Deletion of the CBS Domain from Bacillus anthracis str. Ames complexed with inhibitor D67
To be Published, 2014
|
|
4JHX
 
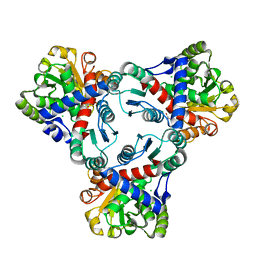 | | Crystal structure of anabolic ornithine carbamoyltransferase from Vibrio vulnificus in complex with carbamoylphosphate and arginine | | Descriptor: | ARGININE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Bacal, P, Winsor, J, Grimshaw, S, Grabowski, M, Chordia, M.D, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-05 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures and kinetic properties of anabolic ornithine carbamoyltransferase from human pathogens Vibrio vulnificus and Bacillus anthracis
To be Published
|
|
4R9X
 
 | | Crystal Structure of Putative Copper Homeostasis Protein CutC from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Copper homeostasis protein CutC, ... | | Authors: | Kim, Y, Zhou, M, Makowska-Grzyska, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-08 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8515 Å) | | Cite: | Crystal Structure of Putative Copper Homeostasis Protein CutC
from Bacillus anthracis
To be Published, 2014
|
|
3UHF
 
 | | Crystal Structure of Glutamate Racemase from Campylobacter jejuni subsp. jejuni | | Descriptor: | CHLORIDE ION, D-GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Maltseva, N, Mulligan, R, Kwon, K, Kim, Y, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-03 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Glutamate Racemase
from Campylobacter jejuni subsp. jejuni
To be Published
|
|
4R7J
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase with the Internal Deletion Containing CBS Domain from Campylobacter jejuni | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1172 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase with the Internal Deletion Containing CBS Domain from Campylobacter jejuni
To be Published, 2014
|
|
4HN3
 
 | | The crystal structure of a sex pheromone precursor (lmo1757) from Listeria monocytogenes EGD-e | | Descriptor: | BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-10-18 | | Release date: | 2012-10-31 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | The crystal structure of a sex pheromone precursor (lmo1757) from Listeria monocytogenes EGD-e
To be Published
|
|
4E5S
 
 | |
3UXJ
 
 | | Crystal Structure of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with NADP and PreQ0 | | Descriptor: | 1,2-ETHANEDIOL, 7-DEAZA-7-AMINOMETHYL-GUANINE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kim, Y, Zhang, R, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-05 | | Release date: | 2012-01-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal Structure of 7-cyano-7-deazaguanine reductase, QueF from
Vibrio cholerae complexed with NADP and PreQ0
To be Published, 2012
|
|
7K9K
 
 | |
7K9I
 
 | |
7K9H
 
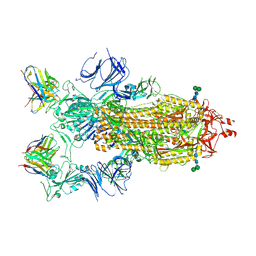 | | SARS-CoV-2 Spike in complex with neutralizing Fab 2B04 (one up, two down conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B04 heavy chain, ... | | Authors: | Errico, J.M, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of SARS-CoV-2 neutralization by two murine antibodies targeting the RBD.
Cell Rep, 37, 2021
|
|
7K9J
 
 | |
4MFG
 
 | | 2.0 Angstrom Resolution Crystal Structure of Putative Carbonic Anhydrase from Clostridium difficile. | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, Putative acyltransferase | | Authors: | Minasov, G, Wawrzak, Z, Kudritska, M, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of Putative Carbonic Anhydrase from Clostridium difficile.
TO BE PUBLISHED
|
|
4M9U
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
4MAM
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ADP analog, AMP-CP | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Phosphoribosylaminoimidazole carboxylase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-16 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ADP analog, AMP-CP
To be Published
|
|
4ECM
 
 | | 2.3 Angstrom Crystal Structure of a Glucose-1-phosphate Thymidylyltransferase from Bacillus anthracis in Complex with Thymidine-5-diphospho-alpha-D-glucose and Pyrophosphate | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, Glucose-1-phosphate thymidylyltransferase, PYROPHOSPHATE 2- | | Authors: | Minasov, G, Kuhn, M, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme glucose-1-phosphate thymidylyltransferase (RfbA).
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4MGE
 
 | | 1.85 Angstrom Resolution Crystal Structure of PTS System Cellobiose-specific Transporter Subunit IIB from Bacillus anthracis. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PTS system, ... | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Winsor, J, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-28 | | Release date: | 2013-09-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of PTS System Cellobiose-specific Transporter Subunit IIB from Bacillus anthracis.
TO BE PUBLISHED
|
|
7KYE
 
 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with CHES | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetyltransferase PA3944, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with CHES
To Be Published
|
|
7KYU
 
 | | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1H-indole-5-carbonyl)oxy]-1H-benzotriazole, 3C-like proteinase | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate
To Be Published
|
|
7KZW
 
 | | Crystal structure of FTT_1639c from Francisella tularensis str. tularensis SCHU S4 | | Descriptor: | CHLORIDE ION, FTT_1639c | | Authors: | Stogios, P.J, Skarina, T, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-10 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of FTT_1639c from Francisella tularensis str. tularensis SCHU S4
To Be Published
|
|
4ECD
 
 | | 2.5 Angstrom Resolution Crystal Structure of Bifidobacterium longum Chorismate Synthase | | Descriptor: | CHLORIDE ION, Chorismate synthase | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2.5 Angstrom Resolution Crystal Structure of Bifidobacterium longum Chorismate Synthase
TO BE PUBLISHED
|
|
4MI1
 
 | | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with aspartyl sulfamoyl adenylates | | Descriptor: | 5'-O-(L-alpha-aspartylsulfamoyl)adenosine, Microcin immunity protein MccF, SULFATE ION | | Authors: | Nocek, B, Severinov, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-30 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the double mutant (S112A, H303A) of B.anthracis mycrocine immunity protein (MccF) with aspartyl sulfamoyl adenylates
TO BE PUBLISHED
|
|
