8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD4
 
 | | PI 3-kinase alpha with nanobody 3-142 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCX
 
 | | PI 3-kinase alpha with nanobody 3-159 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
9J1P
 
 | | Cryo-EM structure of the g1:Ox-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Fan, S, Li, J, Zhuang, J, Zhou, Q, Mai, Y, Lin, B, Wang, M.-W, Wu, C. | | Deposit date: | 2024-08-05 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide-Directed Multicyclic Peptides with N-Terminally Extendable alpha-Helices for Recognition and Activation of G Protein-Coupled Receptors.
J.Am.Chem.Soc., 147, 2025
|
|
9JVX
 
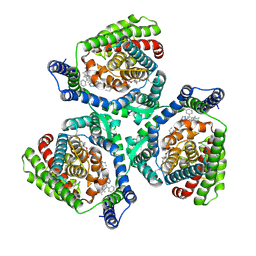 | | Overall structure of human EAAT2 bound with activator (GT949) | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 3-[(~{R})-(4-cyclohexylpiperazin-1-yl)-[1-(2-phenylethyl)-1,2,3,4-tetrazol-5-yl]methyl]-6-methoxy-1~{H}-quinolin-2-one, CHOLESTEROL, ... | | Authors: | Xia, L.Y, Zhang, Y.Y, Shi, Y, Huang, J, Zhou, Q. | | Deposit date: | 2024-10-09 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural study of human glutamate transporter EAAT2
Dian Zi Xian Wei Xue Bao, 2025
|
|
9JVW
 
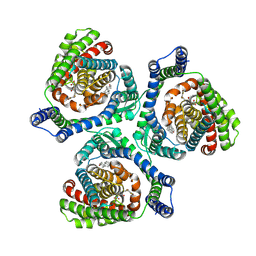 | | Overall structure of human EAAT2 bound with inhibitor (WAY-213613) | | Descriptor: | (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]amino]-4-oxidanylidene-butanoic acid, CHOLESTEROL, Excitatory amino acid transporter 2 | | Authors: | Xia, L.Y, Zhang, Y.Y, Shi, Y, Huang, J, Zhou, Q. | | Deposit date: | 2024-10-09 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural study of human glutamate transporter EAAT2
Dian Zi Xian Wei Xue Bao, 2025
|
|
9JVV
 
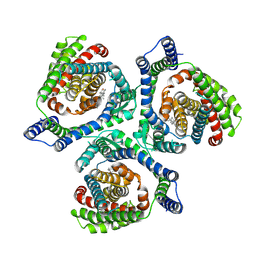 | | Overall structure of human EAAT2 in the substrate-free state | | Descriptor: | CHOLESTEROL, Excitatory amino acid transporter 2 | | Authors: | Xia, L.Y, Zhang, Y.Y, Shi, Y, Huang, J, Zhou, Q. | | Deposit date: | 2024-10-09 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural study of human glutamate transporter EAAT2
Dian Zi Xian Wei Xue Bao, 2025
|
|
5VBL
 
 | | Structure of apelin receptor in complex with agonist peptide | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apelin receptor,Rubredoxin,Apelin receptor Chimera, ZINC ION, ... | | Authors: | Ma, Y, Yue, Y, Ma, Y, Zhang, Q, Zhou, Q, Song, Y, Shen, Y, Li, X, Ma, X, Li, C, Hanson, M.A, Han, G.W, Sickmier, E.A, Swaminath, G, Zhao, S, Stevems, R.C, Hu, L.A, Zhong, W, Zhang, M, Xu, F. | | Deposit date: | 2017-03-29 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Apelin Control of the Human Apelin Receptor
Structure, 25, 2017
|
|
6A69
 
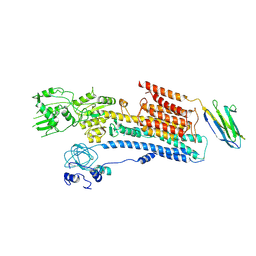 | | Cryo-EM structure of a P-type ATPase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroplastin, Plasma membrane calcium-transporting ATPase 1 | | Authors: | Gong, D.S, Chi, X.M, Ren, K, Huang, G.X.Y, Zhou, G.W, Yan, N, Lei, J.L, Zhou, Q. | | Deposit date: | 2018-06-27 | | Release date: | 2018-09-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Structure of the human plasma membrane Ca2+-ATPase 1 in complex with its obligatory subunit neuroplastin.
Nat Commun, 9, 2018
|
|
6JH6
 
 | | Structure of RyR2 (F/A/Ca2+ dataset) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Peptidyl-prolyl cis-trans isomerase FKBP1B, RyR2, ... | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JGZ
 
 | | Structure of RyR2 (F/P/Ca2+ dataset) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, RyR2, ZINC ION | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-16 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6JG3
 
 | | Cryo-EM structure of RyR2 (Ca2+ alone dataset) | | Descriptor: | Ryanodine receptor 2, ZINC ION | | Authors: | Chi, X.M, Gong, D.S, Ren, K, Zhou, G.W, Huang, G.X.Y, Lei, J.L, Zhou, Q, Yan, N. | | Deposit date: | 2019-02-13 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8XL2
 
 | | Human acetyl-CoA carboxylase 1 filament in complex with acetyl-CoA (ACC1-inact) | | Descriptor: | ACETYL COENZYME *A, Acetyl-CoA carboxylase 1 | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase.
Sci Adv, 10, 2024
|
|
8XL1
 
 | | Core region of the human acetyl-CoA carboxylase 1 filament in complex with acetyl-CoA (ACC1-inact) | | Descriptor: | ACETYL COENZYME *A, Acetyl-CoA carboxylase 1 | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase.
Sci Adv, 10, 2024
|
|
8XL8
 
 | | Structure of human 3-methylcrotonyl-CoA carboxylase in complex with propionyl-CoA (MCC-PCO) | | Descriptor: | BIOTIN, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
8XL0
 
 | | Citrate-induced filament of human acetyl-coenzyme A carboxylase 1 (ACC1-citrate) | | Descriptor: | Acetyl-CoA carboxylase 1, BIOTIN | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase.
Sci Adv, 10, 2024
|
|
8XKZ
 
 | | Core region of the citrate-induced human acetyl-CoA carboxylase 1 filament (ACC1-citrate) | | Descriptor: | Acetyl-CoA carboxylase 1, BIOTIN | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Filament structures unveil the dynamic organization of human acetyl-CoA carboxylase.
Sci Adv, 10, 2024
|
|
8XL3
 
 | | Structure of human propionyl-CoA carboxylase at apo-state (PCC-Apo) | | Descriptor: | BIOTIN, Propionyl-CoA carboxylase alpha chain, mitochondrial, ... | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
8XL6
 
 | | Structure of human 3-methylcrotonyl-CoA carboxylase at apo-state (MCC-Apo) | | Descriptor: | BIOTIN, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
8XL5
 
 | | Structure of human propionyl-CoA carboxylase in complex with propionyl-CoA (PCC-PCO) | | Descriptor: | BIOTIN, Propionyl-CoA carboxylase alpha chain, mitochondrial, ... | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
8XL4
 
 | | Structure of human propionyl-CoA carboxylase in complex with acetyl-CoA (PCC-ACO) | | Descriptor: | ACETYL COENZYME *A, BIOTIN, Propionyl-CoA carboxylase alpha chain, ... | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
8XL7
 
 | | Structure of human 3-methylcrotonyl-CoA carboxylase in complex with acetyl-CoA (MCC-ACO) | | Descriptor: | ACETYL COENZYME *A, BIOTIN, Methylcrotonoyl-CoA carboxylase beta chain, ... | | Authors: | Zhou, F.Y, Zhang, Y.Y, Zhou, Q, Hu, Q. | | Deposit date: | 2023-12-25 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural insights into human propionyl-CoA carboxylase (PCC) and 3-methylcrotonyl-CoA carboxylase (MCC)
Elife, 2024
|
|
8WYI
 
 | | T cell receptor delta 2 gamma 9 with TCRD TM domain chimera of TRAC | | Descriptor: | Signal peptide,flag tag,T cell receptor delta variable 2,T cell receptor delta constant,T cell receptor alpha chain constant,T cell receptor delta variable 2,T cell receptor delta constant,T cell receptor alpha chain constant, Signal peptide,flag tag,T cell receptor gamma variable 9,T cell receptor gamma constant 1,T cell receptor gamma variable 9,T cell receptor gamma constant 1, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Xin, W, Huang, B, Chi, X, Liu, Y, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8WY0
 
 | | T cell receptor delta 2 gamma 9 with F283A, F290A, and F291A | | Descriptor: | CHOLESTEROL, Signal peptide,flag tag,T cell receptor delta variable 2,T cell receptor delta constant, Signal peptide,flag tag,T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | Authors: | Xin, W, Huang, B, Chi, X, Liu, Y, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
