6AJ9
 
 | | The structure of Enterovirus D68 mature virion in complex with Fab 15C5 and 11G1 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-27 | | Release date: | 2018-11-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6ACW
 
 | | The structure of CVA10 virus procapsid particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Zhu, R, Xu, L.F, Zheng, Q.B, Cui, Y.X, Li, S.W, Yan, X.D, Zhou, Z.H, Cheng, T. | | Deposit date: | 2018-07-27 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10.
Sci Adv, 4, 2018
|
|
5Y9F
 
 | |
6LAT
 
 | | The cryo-EM structure of HEV VLP | | Descriptor: | Protein ORF2 | | Authors: | Zheng, Q, He, M, Li, S. | | Deposit date: | 2019-11-13 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Viral neutralization by antibody-imposed physical disruption.
Proc.Natl.Acad.Sci.USA, 2019
|
|
5XS5
 
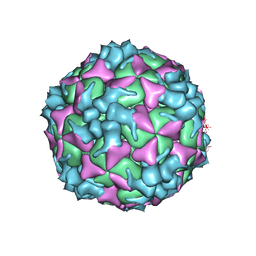 | | Structure of Coxsackievirus A6 (CVA6) virus procapsid particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Cheng, T, Li, S.W. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
6LHB
 
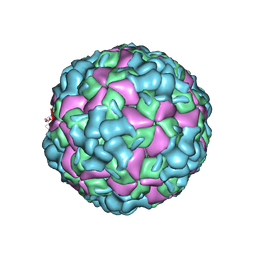 | | The cryo-EM structure of coxsackievirus A16 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHC
 
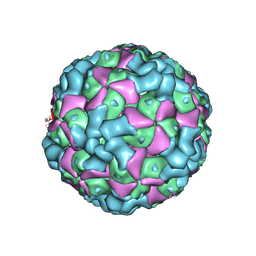 | | The cryo-EM structure of coxsackievirus A16 empty particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHL
 
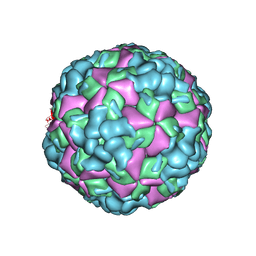 | | The cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab 18A7 | | Descriptor: | VP1 protein, VP2 protein, VP3 protein | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6ACZ
 
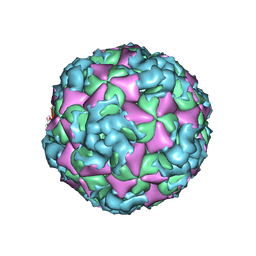 | | The structure of CVA10 virus A-particle from its complex with Fab 2G8 | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, R, Zheng, Q.B, Xu, L.F, Cui, Y.X, Li, S.W, Yan, X.D, Zhou, Z.H, Cheng, T. | | Deposit date: | 2018-07-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10.
Sci Adv, 4, 2018
|
|
6AJ7
 
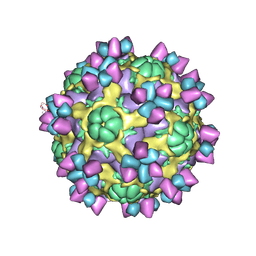 | | The structure of Enterovirus D68 mature virion in complex with Fab 15C5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-27 | | Release date: | 2018-11-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ3
 
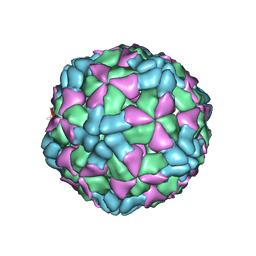 | | The structure of Enterovirus D68 procapsid | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6LHP
 
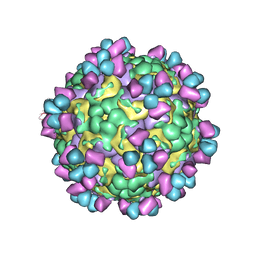 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 14B10 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6ACY
 
 | | The structure of CVA10 virus A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Cui, Y.X, Zheng, Q.B, Zhu, R, Xu, L.F, Li, S.W, Yan, X.D, Zhou, Z.H, Cheng, T. | | Deposit date: | 2018-07-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery and structural characterization of a therapeutic antibody against coxsackievirus A10.
Sci Adv, 4, 2018
|
|
6LHO
 
 | | The cryo-EM structure of coxsackievirus A16 empty particle in complex with Fab 18A7 | | Descriptor: | VP1 protein, VP2 protein, VP3 protein | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
7DNH
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 2H3 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of 2H3 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNK
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 5G9 | | Descriptor: | Major capsid protein L1, The heavy chain of 5G9 Fab fragment, The light chain of 5G9 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (6.41 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNL
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of A4B4 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of A4B4 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
8IX3
 
 | | Cryo-EM structure of SARS-CoV-2 BA.4/5 spike protein in complex with 1G11 (local refinement) | | Descriptor: | BA.4/5 variant spike protein, heavy chain of 1G11, light chain of 1G11 | | Authors: | Sun, H, Jiang, Y, Zheng, Z, Zheng, Q, Li, S. | | Deposit date: | 2023-03-31 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis for broad neutralization of human antibody against Omicron sublineages and evasion by XBB variant.
J.Virol., 97, 2023
|
|
7WI0
 
 | | SARS-CoV-2 Omicron variant spike in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7WHZ
 
 | | SARS-CoV-2 spike protein in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7MF1
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody 47D1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 47D1 Fab heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-04-08 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Diverse immunoglobulin gene usage and convergent epitope targeting in neutralizing antibody responses to SARS-CoV-2.
Cell Rep, 35, 2021
|
|
7EAN
 
 | | immune complex of SARS-CoV-2 RBD and cross-neutralizing antibody 6D6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SARS-CoV-2 cross-neutralizing mAb 6D6, Light chain of SARS-CoV-2 cross-neutralizing mAb 6D6, ... | | Authors: | Li, T.T, Gu, Y, Li, S.W. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants.
Nat Commun, 12, 2021
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | Descriptor: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPF
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion | | Descriptor: | Capsid protein VP4, PALMITIC ACID, VP2, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
