8B56
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the inhibitor GD-9 | | Descriptor: | (2~{S})-4-(2-chloranylethanoyl)-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5, BROMIDE ION, ... | | Authors: | Straeter, N, Muller, C.E, Claff, T, Sylvester, K, Weisse, R, Gao, S, Song, L, Liu, X, Zhan, P. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Discovery and Crystallographic Studies of Nonpeptidic Piperazine Derivatives as Covalent SARS-CoV-2 Main Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
1B25
 
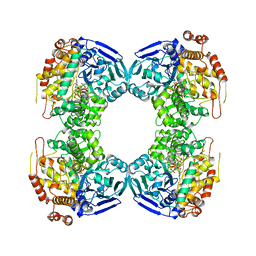 | | FORMALDEHYDE FERREDOXIN OXIDOREDUCTASE FROM PYROCOCCUS FURIOSUS | | Descriptor: | IRON/SULFUR CLUSTER, PROTEIN (FORMALDEHYDE FERREDOXIN OXIDOREDUCTASE), TUNGSTOPTERIN | | Authors: | Hu, Y.L, Faham, S, Roy, R, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1998-12-04 | | Release date: | 1999-03-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Formaldehyde ferredoxin oxidoreductase from Pyrococcus furiosus: the 1.85 A resolution crystal structure and its mechanistic implications.
J.Mol.Biol., 286, 1999
|
|
1B4N
 
 | | FORMALDEHYDE FERREDOXIN OXIDOREDUCTASE FROM PYROCOCCUS FURIOSUS, COMPLEXED WITH GLUTARATE | | Descriptor: | CALCIUM ION, FORMALDEHYDE FERREDOXIN OXIDOREDUCTASE, GLUTARIC ACID, ... | | Authors: | Hu, Y.L, Faham, S, Roy, R, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1998-12-24 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Formaldehyde ferredoxin oxidoreductase from Pyrococcus furiosus: the 1.85 A resolution crystal structure and its mechanistic implications.
J.Mol.Biol., 286, 1999
|
|
3SO8
 
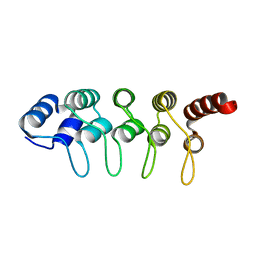 | | Crystal Structure of ANKRA | | Descriptor: | Ankyrin repeat family A protein 2 | | Authors: | Xu, C, Bochkarev, A, Bian, C.B, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-30 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sequence-Specific Recognition of a PxLPxI/L Motif by an Ankyrin Repeat Tumbler Lock.
Sci.Signal., 5, 2012
|
|
3TRA
 
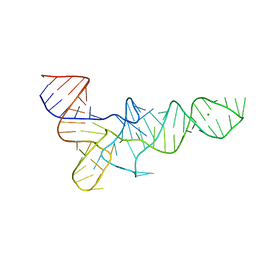 | |
1A1D
 
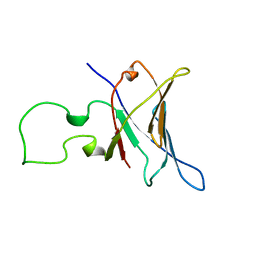 | | YEAST RNA POLYMERASE SUBUNIT RPB8, NMR, MINIMIZED AVERAGE STRUCTURE, ALPHA CARBONS ONLY | | Descriptor: | RNA POLYMERASE | | Authors: | Krapp, S, Kelly, G, Reischl, J, Weinzierl, R, Matthews, S. | | Deposit date: | 1997-12-10 | | Release date: | 1999-03-02 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Eukaryotic RNA polymerase subunit RPB8 is a new relative of the OB family.
Nat.Struct.Biol., 5, 1998
|
|
1HMJ
 
 | | SOLUTION STRUCTURE OF RNA POLYMERASE SUBUNIT H | | Descriptor: | PROTEIN (SUBUNIT H) | | Authors: | Thiru, A, Hodach, M, Eloranta, J, Kostourou, V, Weinzierl, R. | | Deposit date: | 1999-02-05 | | Release date: | 1999-04-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | RNA polymerase subunit H features a beta-ribbon motif within a novel fold that is present in archaea and eukaryotes.
J.Mol.Biol., 287, 1999
|
|
3M10
 
 | | Substrate-free form of Arginine Kinase | | Descriptor: | Arginine kinase, SULFATE ION | | Authors: | Yousef, M.S, Clark, S.A, Pruett, P.K, Somasundaram, T, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.727 Å) | | Cite: | Arginine kinase: joint crystallographic and NMR RDC analyses link substrate-associated motions to intrinsic flexibility.
J.Mol.Biol., 405, 2011
|
|
8VKA
 
 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 9c | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fenwick, M.K, Staker, B.L, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Exploring Subsite Selectivity within Plasmodium vivax N -Myristoyltransferase Using Pyrazole-Derived Inhibitors.
J.Med.Chem., 67, 2024
|
|
8VKB
 
 | | Crystal structure of Plasmodium vivax glycylpeptide N-tetradecanoyltransferase (N-myristoyltransferase, NMT) bound to myristoyl-CoA and inhibitor 10b | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, ... | | Authors: | Fenwick, M.K, Staker, B.L, Phan, I.Q, Early, J, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Exploring Subsite Selectivity within Plasmodium vivax N -Myristoyltransferase Using Pyrazole-Derived Inhibitors.
J.Med.Chem., 67, 2024
|
|
6VZZ
 
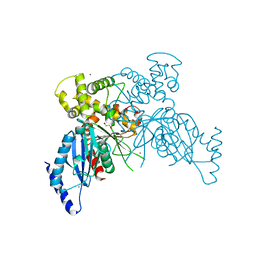 | |
7NDX
 
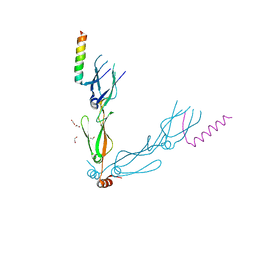 | | Crystal structure of the human HSP40 DNAJB1-CTDs in complex with a peptide of NudC | | Descriptor: | 1,2-ETHANEDIOL, DnaJ homolog subfamily B member 1, Nuclear migration protein nudC | | Authors: | Delhommel, F, Zak, K.M, Popowicz, G.M, Sattler, M. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | NudC guides client transfer between the Hsp40/70 and Hsp90 chaperone systems.
Mol.Cell, 82, 2022
|
|
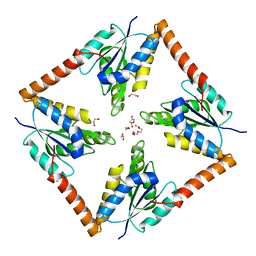
6O55
 
 | |
8TA0
 
 | |
8TBR
 
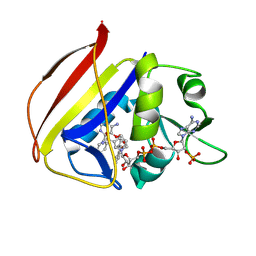 | |
8TA1
 
 | |
5VCJ
 
 | | Structure of alpha-galactosylphytosphingosine bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | (2S,3S,4R)-2-amino-3,4-dihydroxyoctadecyl alpha-D-galactopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wang, J, Zajonc, D.M. | | Deposit date: | 2017-03-31 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Enhancing T cell responses and tumour immunity by vaccination with peptides conjugated to a weak NKT cell agonist.
Org. Biomol. Chem., 17, 2019
|
|
7DM2
 
 | | crystal structure of the M. tuberculosis phosphate ABC transport receptor PstS-1 in complex with Fab p4-170 | | Descriptor: | PHOSPHATE ION, Phosphate-binding protein PstS 1, heavy chain, ... | | Authors: | Ma, B, Freund, N, Xiang, Y. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human antibodies targeting a Mycobacterium transporter protein mediate protection against tuberculosis.
Nat Commun, 12, 2021
|
|
7DM1
 
 | | crystal structure of the M.tuberculosis phosphate ABC transport receptor PstS-1 in complex with Fab p4-36 | | Descriptor: | PHOSPHATE ION, Phosphate-binding protein PstS 1, heavy chain, ... | | Authors: | Ma, B, Freund, N, Xiang, Y. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human antibodies targeting a Mycobacterium transporter protein mediate protection against tuberculosis.
Nat Commun, 12, 2021
|
|
4LG6
 
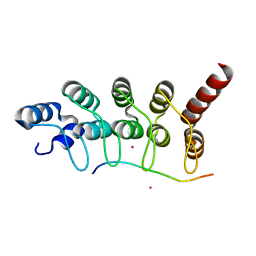 | | Crystal structure of ANKRA2-CCDC8 complex | | Descriptor: | Ankyrin repeat family A protein 2, Coiled-coil domain-containing protein 8, UNKNOWN ATOM OR ION | | Authors: | Xu, C, Bian, C, Tempel, W, Mackenzie, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ankyrin Repeats of ANKRA2 Recognize a PxLPxL Motif on the 3M Syndrome Protein CCDC8.
Structure, 23, 2015
|
|
1I38
 
 | |
1I37
 
 | |
4GRD
 
 | |
6SAZ
 
 | | Cleaved human fetuin-b in complex with crayfish astacin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Astacin, ... | | Authors: | Gomis-Ruth, F.X, Guevara, T, Cuppari, A, Korschgen, H, Schmitz, C, Kuske, M, Yiallouros, I, Floehr, J, Jahnen-Dechent, W, Stocker, W. | | Deposit date: | 2019-07-18 | | Release date: | 2019-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The C-terminal region of human plasma fetuin-B is dispensable for the raised-elephant-trunk mechanism of inhibition of astacin metallopeptidases.
Sci Rep, 9, 2019
|
|
2TRA
 
 | | RESTRAINED REFINEMENT OF TWO CRYSTALLINE FORMS OF YEAST ASPARTIC ACID AND PHENYLALANINE TRANSFER RNA CRYSTALS | | Descriptor: | MAGNESIUM ION, SPERMINE, TRNAASP | | Authors: | Westhof, E, Dumas, P, Moras, D. | | Deposit date: | 1987-11-06 | | Release date: | 1987-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalanine transfer RNA crystals.
Acta Crystallogr.,Sect.A, 44, 1988
|
|
