2ZWA
 
 | | Crystal structure of tRNA wybutosine synthesizing enzyme TYW4 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Leucine carboxyl methyltransferase 2, ... | | Authors: | Suzuki, Y, Noma, A, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2008-12-01 | | Release date: | 2009-06-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of tRNA modification with CO2 fixation and methylation by wybutosine synthesizing enzyme TYW4.
Nucleic Acids Res., 37, 2009
|
|
7BPG
 
 | | Structure of serinol nucleic acid - RNA complex | | Descriptor: | CALCIUM ION, RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3'), SNA (S-(F7R)(F7X)(F7O)(F7R)(F7X)(F7O)(F7R)(F7U)-R) | | Authors: | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | Deposit date: | 2020-03-22 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
7BPF
 
 | | Structure of L-threoninol nucleic acid - RNA complex | | Descriptor: | L-aTNA (3'-(*GP*CP*AP*GP*CP*AP*GP*C)-1'), RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3') | | Authors: | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | Deposit date: | 2020-03-22 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
3VUO
 
 | | Crystal structure of nontoxic nonhemagglutinin subcomponent (NTNHA) from clostridium botulinum serotype D strain 4947 | | Descriptor: | NTNHA | | Authors: | Sagane, Y, Miyashita, S.-I, Miyata, K, Matsumoto, T, Inui, K, Hayashi, S, Suzuki, T, Hasegawa, K, Yajima, S, Yamano, A, Niwa, K, Watanabe, T. | | Deposit date: | 2012-07-03 | | Release date: | 2012-09-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Small-angle X-ray scattering reveals structural dynamics of the botulinum neurotoxin associating protein, nontoxic nonhemagglutinin
Biochem.Biophys.Res.Commun., 425, 2012
|
|
7DUA
 
 | |
7DU9
 
 | |
7DU8
 
 | |
3VUN
 
 | | Crystal structure of a influenza A virus (A/Aichi/2/1968 H3N2) hemagglutinin in C2 space group. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Yasutake, Y, Suzuki, T, Kawaguchi, A, Nobusawa, E. | | Deposit date: | 2012-07-02 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a influenza A virus (A/Aichi/2/1968 H3N2) hemagglutinin in C2 space group
To be Published
|
|
7F5S
 
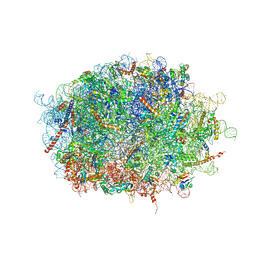 | | human delta-METTL18 60S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Takahashi, M, Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | METTL18-mediated histidine methylation of RPL3 modulates translation elongation for proteostasis maintenance.
Elife, 11, 2022
|
|
4V7N
 
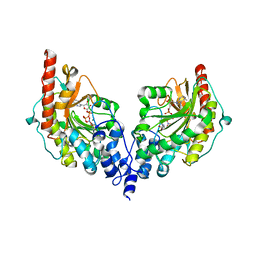 | | Glycocyamine kinase, beta-beta homodimer from marine worm Namalycastis sp., with transition state analog Mg(II)-ADP-NO3-glycocyamine. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANIDINO ACETATE, Glycocyamine kinase beta chain, ... | | Authors: | Lim, K, Pullalarevu, S, Herzberg, O. | | Deposit date: | 2009-12-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the mechanism and substrate specificity of glycocyamine kinase, a phosphagen kinase family member.
Biochemistry, 49, 2010
|
|
8DOX
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-14 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8DPR
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with inhibitor TKB-248 | | Descriptor: | 2,2,2-trifluoro-N-{(2S)-1-[(1R,2S,5S)-2-({(2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamothioyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-3-yl]-3,3-dimethyl-1-oxobutan-2-yl}acetamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7QVP
 
 | | Human collided disome (di-ribosome) stalled on XBP1 mRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Denk, T.G, Tesina, P, Beckmann, R. | | Deposit date: | 2022-01-22 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A distinct mammalian disome collision interface harbors K63-linked polyubiquitination of uS10 to trigger hRQT-mediated subunit dissociation.
Nat Commun, 13, 2022
|
|
3S7T
 
 | |
3L2D
 
 | |
3L2E
 
 | | Glycocyamine kinase, alpha-beta heterodimer from marine worm Namalycastis sp. | | Descriptor: | Glycocyamine kinase alpha chain, Glycocyamine kinase beta chain | | Authors: | Lim, K, Pullalarevu, S, Herzberg, O. | | Deposit date: | 2009-12-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the mechanism and substrate specificity of glycocyamine kinase, a phosphagen kinase family member.
Biochemistry, 49, 2010
|
|
6HSG
 
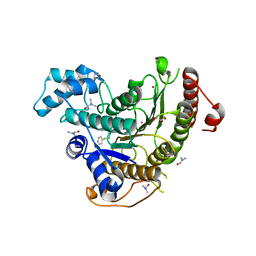 | | Crystal structure of Schistosoma mansoni HDAC8 H292M mutant complexed with NCC-149 | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, Histone deacetylase, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
8GZC
 
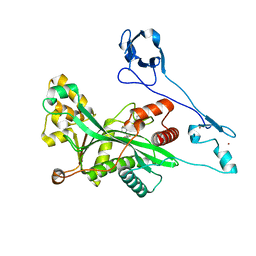 | | Crystal structure of EP300 HAT domain in complex with compound 10 | | Descriptor: | (2~{R},4~{R})-4-fluoranyl-1-[1-(4-methoxyphenyl)cyclohexyl]carbonyl-~{N}-(1~{H}-pyrazolo[4,3-b]pyridin-5-yl)pyrrolidine-2-carboxamide, Histone acetyltransferase p300, ZINC ION | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2022-09-26 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of DS-9300: A Highly Potent, Selective, and Once-Daily Oral EP300/CBP Histone Acetyltransferase Inhibitor.
J.Med.Chem., 66, 2023
|
|
3SIX
 
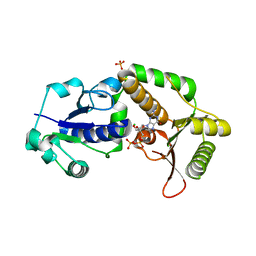 | | Crystal structure of NodZ alpha-1,6-fucosyltransferase soaked with GDP-fucose | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Nodulation fucosyltransferase NodZ, ... | | Authors: | Brzezinski, K, Dauter, Z, Jaskolski, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of NodZ alpha-1,6-fucosyltransferase in complex with GDP and GDP-fucose
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SIW
 
 | | Crystal structure of NodZ alpha-1,6-fucosyltransferase co-crystallized with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Nodulation fucosyltransferase NodZ, PHOSPHATE ION | | Authors: | Brzezinski, K, Dauter, Z, Jaskolski, M. | | Deposit date: | 2011-06-20 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of NodZ alpha-1,6-fucosyltransferase in complex with GDP and GDP-fucose
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5XVR
 
 | |
5XI8
 
 | | Structure and function of the TPR domain | | Descriptor: | Beta-barrel assembly-enhancing protease | | Authors: | Tanaka, Y, Tsukazaki, T. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The TPR domain of BepA is required for productive interaction with substrate proteins and the beta-barrel assembly machinery complex.
Mol. Microbiol., 106, 2017
|
|
7F4V
 
 | | Cryo-EM structure of a primordial cyanobacterial photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Hamaguchi, T, Nagao, R, Kawakami, K, Yonekura, K, Shen, J.R. | | Deposit date: | 2021-06-21 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Structural basis for the absence of low-energy chlorophylls responsible for photoprotection from a primitive cyanobacterial PSI
Biorxiv, 2022
|
|
7VHZ
 
 | | Crystal structure of EP300 HAT domain in complex with compound 7 | | Descriptor: | (2R)-N-(2H-indazol-4-yl)-1-[1-(4-methoxyphenyl)cyclopentyl]carbonyl-pyrrolidine-2-carboxamide, Histone acetyltransferase p300, ZINC ION | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2021-09-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of EP300/CBP histone acetyltransferase inhibitors through scaffold hopping of 1,4-oxazepane ring.
Bioorg.Med.Chem.Lett., 66, 2022
|
|
7VHY
 
 | | Crystal structure of EP300 HAT domain in complex with compound (+)-3 | | Descriptor: | Histone acetyltransferase p300, ZINC ION, [(6R)-6-(1H-indazol-4-ylmethyl)-1,4-oxazepan-4-yl]-[1-(4-methoxyphenyl)cyclopentyl]methanone | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2021-09-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of EP300/CBP histone acetyltransferase inhibitors through scaffold hopping of 1,4-oxazepane ring.
Bioorg.Med.Chem.Lett., 66, 2022
|
|
