8XRT
 
 | | The crystal structure of a GH3 enzyme CcBgl3B | | Descriptor: | CALCIUM ION, GH3 enzyme CcBgl3B | | Authors: | Su, J.Y. | | Deposit date: | 2024-01-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A trapped covalent intermediate as a key catalytic element in the hydrolysis of a GH3 beta-glucosidase: An X-ray crystallographic and biochemical study.
Int.J.Biol.Macromol., 265, 2024
|
|
8XRX
 
 | | The crystal structure of a GH3 enzyme CcBgl3B with glucose and gentiobiose | | Descriptor: | CALCIUM ION, GH3 enzyme CcBgl3B, alpha-D-glucopyranose, ... | | Authors: | Su, J.Y. | | Deposit date: | 2024-01-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A trapped covalent intermediate as a key catalytic element in the hydrolysis of a GH3 beta-glucosidase: An X-ray crystallographic and biochemical study.
Int.J.Biol.Macromol., 265, 2024
|
|
8XRV
 
 | | The crystal structure of a GH3 enzyme CcBgl3B with glucose | | Descriptor: | CALCIUM ION, GH3 enzyme CcBgl3B, beta-D-glucopyranose | | Authors: | Su, J.Y. | | Deposit date: | 2024-01-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A trapped covalent intermediate as a key catalytic element in the hydrolysis of a GH3 beta-glucosidase: An X-ray crystallographic and biochemical study.
Int.J.Biol.Macromol., 265, 2024
|
|
8XRU
 
 | | The crystal structure of a GH3 enzyme CcBgl3B with glycerol | | Descriptor: | CALCIUM ION, GH3 enzyme CcBgl3B, GLYCEROL | | Authors: | Su, J.Y. | | Deposit date: | 2024-01-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A trapped covalent intermediate as a key catalytic element in the hydrolysis of a GH3 beta-glucosidase: An X-ray crystallographic and biochemical study.
Int.J.Biol.Macromol., 265, 2024
|
|
8SIX
 
 | | Structure of Compound 13 bound to the CHK1 10-point mutant | | Descriptor: | (1S)-N-(7-chloro-6-{4-[(3R,4R)-4-hydroxy-3-methyloxolan-3-yl]piperazin-1-yl}isoquinolin-3-yl)-6-oxaspiro[2.5]octane-1-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2023-04-17 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease.
J.Med.Chem., 66, 2023
|
|
8SIW
 
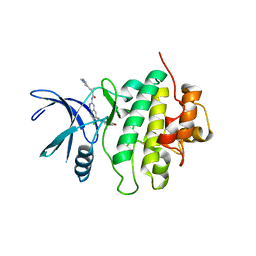 | | Structure of Compound 5 bound to the CHK1 10-point mutant | | Descriptor: | (1S,2S)-N-(7-chloro-6-{1-[(3R,4R)-4-hydroxy-3-methyloxolan-3-yl]piperidin-4-yl}isoquinolin-3-yl)-2-(1-methyl-1H-pyrazol-4-yl)cyclopropane-1-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2023-04-17 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease.
J.Med.Chem., 66, 2023
|
|
8SIV
 
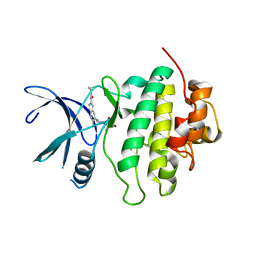 | | Structure of Compound 2 bound to the CHK1 10-point mutant | | Descriptor: | N-(7-chloro-6-{1-[(3R,4R)-4-hydroxy-3-methyloxolan-3-yl]piperidin-4-yl}isoquinolin-3-yl)cyclopropanecarboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2023-04-17 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease.
J.Med.Chem., 66, 2023
|
|
4S0T
 
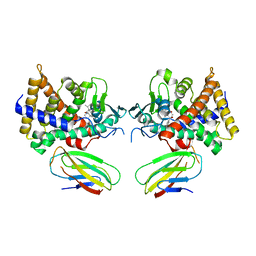 | |
4S0S
 
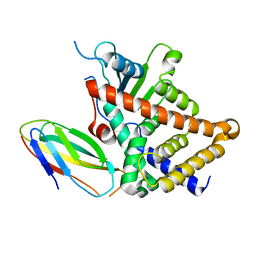 | |
7SUF
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 06 | | Descriptor: | 1,2-ETHANEDIOL, 8-cyclopropyl-N-[5-methyl-1-(oxan-4-yl)-1H-pyrazol-4-yl]quinazolin-2-amine, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7SUJ
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 24 | | Descriptor: | (3R,4R)-4-{4-[6-chloro-2-({1-[(1R)-2,2-difluorocyclopropyl]-5-methyl-1H-pyrazol-4-yl}amino)quinazolin-7-yl]piperidin-1-yl}-4-methyloxolan-3-ol, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7SUH
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 15 | | Descriptor: | 1-[5-chloro-4-({6-chloro-7-[1-(oxetan-3-yl)piperidin-4-yl]quinazolin-2-yl}amino)-1H-pyrazol-1-yl]-2-methylpropan-2-ol, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7SUI
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 22 | | Descriptor: | (3R,4R)-4-{(3S,4S)-4-[6-chloro-2-({5-chloro-1-[(1R)-2,2-difluorocyclopropyl]-1H-pyrazol-4-yl}amino)quinazolin-7-yl]-3-fluoropiperidin-1-yl}oxolan-3-ol, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7SUG
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 09 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-{[5-methyl-1-(oxan-4-yl)-1H-pyrazol-4-yl]amino}quinazolin-8-yl)cyclopropane-1-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
6UQS
 
 | |
6UQT
 
 | |
6UQU
 
 | |
6UR3
 
 | |
5FDP
 
 | | Structure of DDR1 receptor tyrosine kinase in complex with D2099 inhibitor at 2.25 Angstroms resolution. | | Descriptor: | (4~{S})-4-methyl-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinoline-7-carboxamide, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, Arrowsmith, C.H, von Delft, F, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-16 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design of Tetrahydroisoquinoline-7-carboxamides as Selective Discoidin Domain Receptor 1 (DDR1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
7FG9
 
 | | Alpha-1,2-glucosyltransferase_UDP_tll1591 | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Su, J.Y. | | Deposit date: | 2021-07-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for glucosylsucrose synthesis by a member of the alpha-1,2-glucosyltransferase family
Acta Biochim.Biophys.Sin., 54, 2022
|
|
7FGA
 
 | | Alpha-1,2-glucosyltransferase_UDP_sucrose_tll1591 | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE, alpha-D-glucopyranose-(1-1)-alpha-D-fructofuranose | | Authors: | Su, J.Y. | | Deposit date: | 2021-07-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for glucosylsucrose synthesis by a member of the alpha-1,2-glucosyltransferase family
Acta Biochim.Biophys.Sin., 54, 2022
|
|
5GJT
 
 | | Crystal structure of H1 hemagglutinin from A/Washington/05/2011 in complex with a neutralizing antibody 3E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, heavy chain of human neutralizing antibody 3E1, ... | | Authors: | Wang, W, Zhang, T, Ding, J. | | Deposit date: | 2016-07-01 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Human antibody 3E1 targets the HA stem region of H1N1 and H5N6 influenza A viruses
Nat Commun, 7, 2016
|
|
5GJS
 
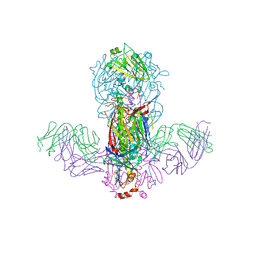 | | Crystal structure of H1 hemagglutinin from A/California/04/2009 in complex with a neutralizing antibody 3E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Wang, W, Zhang, T, Ding, J. | | Deposit date: | 2016-07-01 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Human antibody 3E1 targets the HA stem region of H1N1 and H5N6 influenza A viruses
Nat Commun, 7, 2016
|
|
7UYA
 
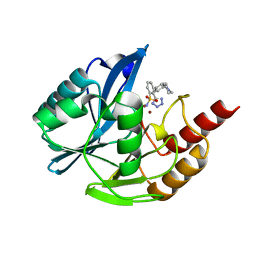 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYD
 
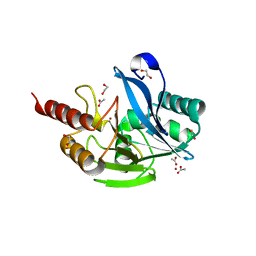 | | Inhibitor bound VIM1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
