1UF9
 
 | | Crystal structure of TT1252 from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, TT1252 protein | | Authors: | Seto, A, Murayama, K, Toyama, M, Ebihara, A, Nakagawa, N, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | ATP-induced structural change of dephosphocoenzyme A kinase from Thermus thermophilus HB8
PROTEINS, 58, 2005
|
|
1UFB
 
 | | Crystal structure of TT1696 from Thermus thermophilus HB8 | | Descriptor: | TT1696 protein | | Authors: | Idaka, M, Murayama, K, Yamaguchi, H, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of TT1696 from Thermus thermophilus HB8
To be published
|
|
1UFA
 
 | | Crystal structure of TT1467 from Thermus thermophilus HB8 | | Descriptor: | TT1467 protein | | Authors: | Idaka, M, Terada, T, Murayama, K, Yamaguchi, H, Nureki, O, Ishitani, R, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-28 | | Release date: | 2003-11-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TT1467 from Thermus thermophilus HB8
To be published
|
|
1UF3
 
 | | Crystal structure of TT1561 of thermus thermophilus HB8 | | Descriptor: | CALCIUM ION, hypothetical protein TT1561 | | Authors: | Kato-Murayama, M, Shirouzu, M, Terada, T, Murayama, K, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-23 | | Release date: | 2003-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of TT1561 of thermus thermophilus HB8
To be Published
|
|
8HAL
 
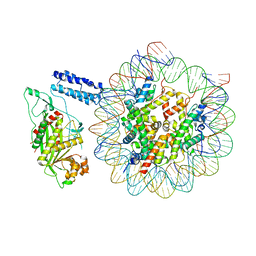 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 1 | | Descriptor: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAG
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 1 (3.2 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAM
 
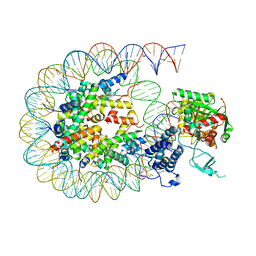 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 2 | | Descriptor: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAH
 
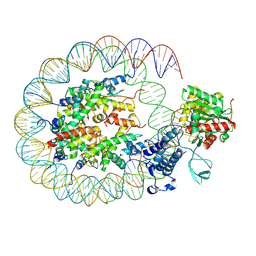 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (3.9 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAI
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 1 (4.7 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAN
 
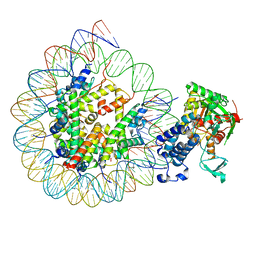 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 3 | | Descriptor: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAJ
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (4.8 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAK
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 4 (4.5 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
5H0B
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanoic acid | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]azaniumyl]-4-methyl-pentanoate, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0H
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-N,N,4-trimethylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-~{N},~{N},4-trimethyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0E
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-4-methylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-4-methyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
5H0G
 
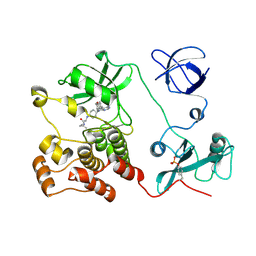 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor (S)-2-(((1r,4S)-4-(4-amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl)cyclohexyl)amino)-N,4-dimethylpentanamide | | Descriptor: | (2~{S})-2-[[4-[4-azanyl-5-(4-phenoxyphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]cyclohexyl]amino]-~{N},4-dimethyl-pentanamide, Tyrosine-protein kinase HCK | | Authors: | Tomabechi, Y, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-10-04 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activity cliff for 7-substituted pyrrolo-pyrimidine inhibitors of HCK explained in terms of predicted basicity of the amine nitrogen.
Bioorg. Med. Chem., 25, 2017
|
|
8Y2O
 
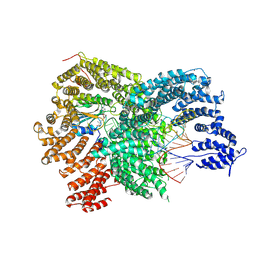 | | The Cryo-EM structure of human tRNA methyltransferase FTSJ1-THADA with substrate tRNA and S-adenosyl homocysteine (SAH) | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Ishiguro, K, Fujimura, A, Shirouzu, M. | | Deposit date: | 2024-01-26 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural insights into tRNA recognition of the human FTSJ1-THADA complex.
Commun Biol, 8, 2025
|
|
7Y6L
 
 | |
7Y6N
 
 | |
1UFM
 
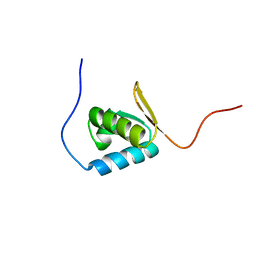 | | Solution structure of the PCI domain | | Descriptor: | COP9 complex subunit 4 | | Authors: | Suzuki, S, Hatanaka, H, Kigawa, T, Shirouzu, M, Hayashizaki, Y, The RIKEN Genome Exploration Research Group Phase I & II Teams, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-02 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PCI domain
To be Published
|
|
8Z7D
 
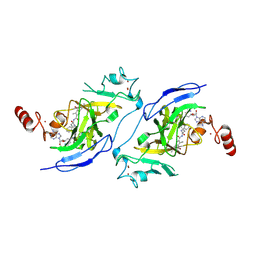 | | Structure of G9a in complex with compound 9a | | Descriptor: | 3,6,6-trimethyl-4-oxidanylidene-~{N}-[(2~{S})-1-oxidanylidene-1-[(phenylmethyl)amino]hexan-2-yl]-5,7-dihydro-1~{H}-indole-2-carboxamide, Histone-lysine N-methyltransferase EHMT2, SINEFUNGIN, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Ihara, K, Shirouzu, M, Umehara, T. | | Deposit date: | 2024-04-20 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Structure-based development of novel substrate-type G9a inhibitors as epigenetic modulators for sickle cell disease treatment.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
8Z7E
 
 | | Structure of G9a in complex with compound 9b | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, SINEFUNGIN, ZINC ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Ihara, K, Shirouzu, M, Umehara, T. | | Deposit date: | 2024-04-20 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.545 Å) | | Cite: | Structure-based development of novel substrate-type G9a inhibitors as epigenetic modulators for sickle cell disease treatment.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
8Z7C
 
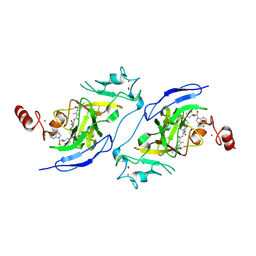 | | Structure of G9a in complex with compound 7i | | Descriptor: | 3,6,6-trimethyl-~{N}-[(2~{S})-1-[[4-(1-methylpiperidin-4-yl)oxyphenyl]amino]-1-oxidanylidene-hexan-2-yl]-4-oxidanylidene-5,7-dihydro-1~{H}-indole-2-carboxamide, Histone-lysine N-methyltransferase EHMT2, SINEFUNGIN, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Ihara, K, Shirouzu, M, Umehara, T. | | Deposit date: | 2024-04-20 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure-based development of novel substrate-type G9a inhibitors as epigenetic modulators for sickle cell disease treatment.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
8K9L
 
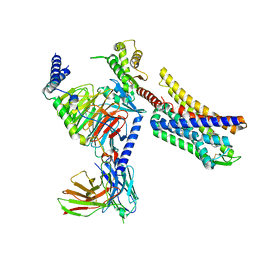 | | Full agonist- and positive allosteric modulator-bound mu-type opioid receptor-G protein complex | | Descriptor: | (2~{S})-2-(3-bromanyl-4-methoxy-phenyl)-3-(4-chlorophenyl)sulfonyl-1,3-thiazolidine, DAMGO, G protein subunit alpha i3, ... | | Authors: | Hisano, T, Uchikubo-Kamo, T, Shirouzu, M, Imai, S, Kaneko, S, Shimada, I. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural and dynamic insights into the activation of the mu-opioid receptor by an allosteric modulator.
Nat Commun, 15, 2024
|
|
8K9K
 
 | | Full agonist-bound mu-type opioid receptor-G protein complex | | Descriptor: | DAMGO, G protein subunit alpha i3, Guanine nucleotide binding protein, ... | | Authors: | Hisano, T, Uchikubo-Kamo, T, Shirouzu, M, Imai, S, Kaneko, S, Shimada, I. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural and dynamic insights into the activation of the mu-opioid receptor by an allosteric modulator.
Nat Commun, 15, 2024
|
|
