1EAF
 
 | |
3ULS
 
 | | Crystal structure of Fab12 | | Descriptor: | Fab12 heavy chain, Fab12 light chain | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
3ULU
 
 | | Structure of quaternary complex of human TLR3ecd with three Fabs (Form1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1068 heavy chain, ... | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
3ULV
 
 | | Structure of quaternary complex of human TLR3ecd with three Fabs (Form2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1068 heavy chain, ... | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.522 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
1DPB
 
 | |
1DPD
 
 | |
1DPC
 
 | |
7JGT
 
 | | Crystal Structure of FN3tt | | Descriptor: | Fibronectin type-III domain-containing protein | | Authors: | Luo, J, Malia, T.J. | | Deposit date: | 2020-07-19 | | Release date: | 2021-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Surface salt bridges contribute to the extreme thermal stability of an FN3-like domain from a thermophilic bacterium.
Proteins, 90, 2022
|
|
7JGU
 
 | | Structure of FN3tt mut | | Descriptor: | Fibronectin type-III domain-containing protein, PENTAETHYLENE GLYCOL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2020-07-19 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Surface salt bridges contribute to the extreme thermal stability of an FN3-like domain from a thermophilic bacterium.
Proteins, 90, 2022
|
|
6CK8
 
 | |
3O8O
 
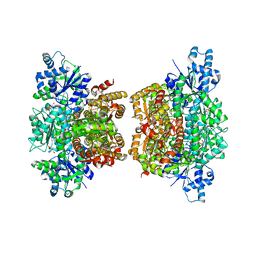 | | Structure of phosphofructokinase from Saccharomyces cerevisiae | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructokinase subunit alpha, ... | | Authors: | Banaszak, K, Mechin, I, Kopperschlager, G, Rypniewski, W. | | Deposit date: | 2010-08-03 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structures of Eukaryotic Phosphofructokinases from Baker's Yeast and Rabbit Skeletal Muscle.
J.Mol.Biol., 407, 2011
|
|
1DEK
 
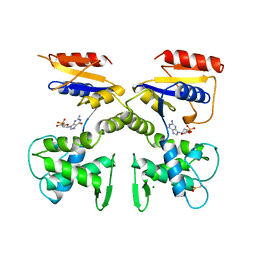 | |
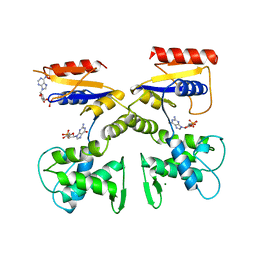
1DEL
 
 | |
3O8N
 
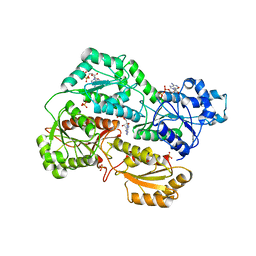 | | Structure of phosphofructokinase from rabbit skeletal muscle | | Descriptor: | 6-phosphofructokinase, muscle type, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Banaszak, K, Chang, S.H, Rypniewski, W. | | Deposit date: | 2010-08-03 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structures of Eukaryotic Phosphofructokinases from Baker's Yeast and Rabbit Skeletal Muscle.
J.Mol.Biol., 407, 2011
|
|
3O8L
 
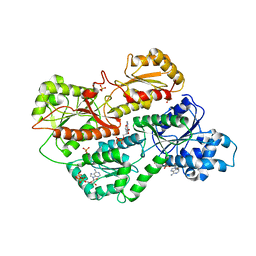 | | Structure of phosphofructokinase from rabbit skeletal muscle | | Descriptor: | 6-phosphofructokinase, muscle type, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Banaszak, K, Chang, S.H, Rypniewski, W. | | Deposit date: | 2010-08-03 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structures of Eukaryotic Phosphofructokinases from Baker's Yeast and Rabbit Skeletal Muscle.
J.Mol.Biol., 407, 2011
|
|
6FYT
 
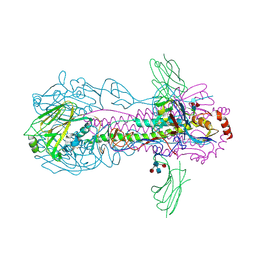 | |
6FYW
 
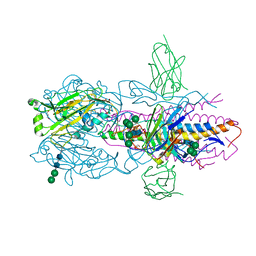 | |
6FYU
 
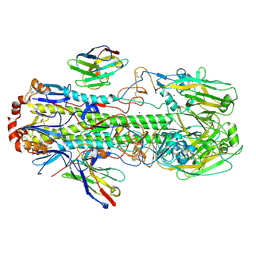 | | Structure of H7(A/Shanghai/2/2013) Influenza Hemagglutinin in complex SD36 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Laursen, N.S, Wilson, I.A. | | Deposit date: | 2018-03-12 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.643 Å) | | Cite: | Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin.
Science, 362, 2018
|
|
6FYS
 
 | | Structure of single domain antibody SD83 | | Descriptor: | 1,2-ETHANEDIOL, Single domain antibody SD83 | | Authors: | Laursen, N.S, Wilson, I.A. | | Deposit date: | 2018-03-12 | | Release date: | 2018-11-07 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin.
Science, 362, 2018
|
|
1BKW
 
 | | p-Hydroxybenzoate hydroxylase (phbh) mutant with cys116 replaced by ser (c116s) and arg44 replaced by lys (r44k), in complex with fad and 4-hydroxybenzoic acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H, Schreuder, H.A, Van Berkel, W.J. | | Deposit date: | 1998-07-13 | | Release date: | 1998-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of mutant Arg44Lys of 4-hydroxybenzoate hydroxylase implications for NADPH binding.
Eur.J.Biochem., 231, 1995
|
|
1CC6
 
 | | PHE161 AND ARG166 VARIANTS OF P-HYDROXYBENZOATE HYDROXYLASE. IMPLICATIONS FOR NADPH RECOGNITION AND STRUCTURAL STABILITY. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Bunthof, C, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-03-04 | | Release date: | 1999-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phe161 and Arg166 variants of p-hydroxybenzoate hydroxylase. Implications for NADPH recognition and structural stability.
Febs Lett., 443, 1999
|
|
1CC4
 
 | | PHE161 AND ARG166 VARIANTS OF P-HYDROXYBENZOATE HYDROXYLASE. IMPLICATIONS FOR NADPH RECOGNITION AND STRUCTURAL STABILITY. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Bunthof, C, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-03-04 | | Release date: | 1999-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phe161 and Arg166 variants of p-hydroxybenzoate hydroxylase. Implications for NADPH recognition and structural stability.
Febs Lett., 443, 1999
|
|
1BGN
 
 | | P-HYDROXYBENZOATE HYDROXYLASE (PHBH) MUTANT WITH CYS 116 REPLACED BY SER (C116S) AND ARG 269 REPLACED BY THR (R269T), IN COMPLEX WITH FAD AND 4-HYDROXYBENZOIC ACID | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Eppink, M.H.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1998-05-29 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interdomain binding of NADPH in p-hydroxybenzoate hydroxylase as suggested by kinetic, crystallographic and modeling studies of histidine 162 and arginine 269 variants.
J.Biol.Chem., 273, 1998
|
|
1CJ4
 
 | | MUTANT Q34T OF PARA-HYDROXYBENZOATE HYDROXYLASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Overkamp, K.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-04-21 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Switch of coenzyme specificity of p-hydroxybenzoate hydroxylase.
J.Mol.Biol., 292, 1999
|
|
1CJ3
 
 | | MUTANT TYR38GLU OF PARA-HYDROXYBENZOATE HYDROXYLASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Overkamp, K.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-04-21 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Switch of coenzyme specificity of p-hydroxybenzoate hydroxylase.
J.Mol.Biol., 292, 1999
|
|
