1I4R
 
 | |
5YJI
 
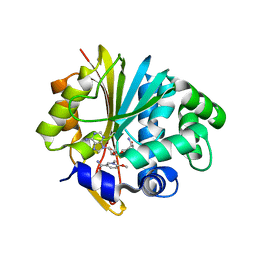 | | Co-crystal structure of Mouse Nicotinamide N-methyltransferase (NNMT) with small molecule analog of Nicotinamide | | Descriptor: | 6-methoxy-1-methyl-2H-pyridine-3-carboxamide, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Birudukota, S, Swaminathan, S, Thakur, M.K, Parveen, R, Kandan, S, Hallur, M.S, Rajagopal, S, Ruf, S, Dhakshinamoorthy, S, Kannt, A, Gosu, R. | | Deposit date: | 2017-10-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A small molecule inhibitor of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 8, 2018
|
|
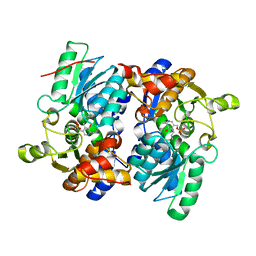
5JIS
 
 | |
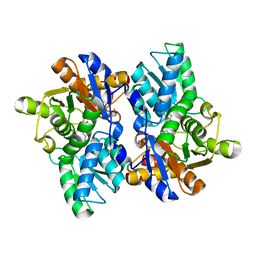
5JJC
 
 | |
5JZX
 
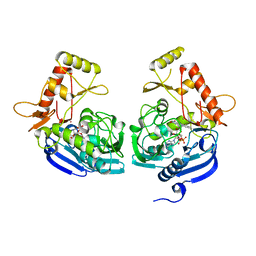 | | Crystal Structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Mycobacterium tuberculosis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, POTASSIUM ION, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Dharavath, S, Eniyan, K, Bajpai, U, Gourinath, S. | | Deposit date: | 2016-05-17 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine-enolpyruvate reductase (MurB) from Mycobacterium tuberculosis
Biochim. Biophys. Acta, 1866, 2017
|
|
2QJC
 
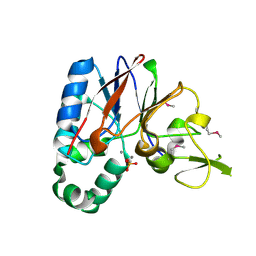 | | Crystal structure of a putative diadenosine tetraphosphatase | | Descriptor: | Diadenosine tetraphosphatase, putative, MANGANESE (II) ION, ... | | Authors: | Sugadev, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-06 | | Release date: | 2007-07-24 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
1NKQ
 
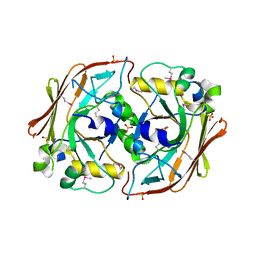 | | Crystal structure of yeast ynq8, a fumarylacetoacetate hydrolase family protein | | Descriptor: | ACETIC ACID, CALCIUM ION, Hypothetical 28.8 kDa protein in PSD1-SKO1 intergenic region, ... | | Authors: | Eswaramoorthy, S, Kumaran, D, Daniels, B, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-03 | | Release date: | 2004-06-15 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crtystal Structure of Yeast Hypothetical Protein YNQ8_YEAST
To be Published
|
|
1NJR
 
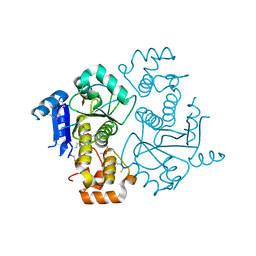 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase | | Descriptor: | 32.1 kDa protein in ADH3-RCA1 intergenic region, Xylitol | | Authors: | Kumaran, D, Eswaramoorthy, S, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-02 | | Release date: | 2004-08-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of ADP-ribose-1''-monophosphatase (Appr-1''-pase), a ubiquitous cellular processing enzyme
Protein Sci., 14, 2005
|
|
5YO2
 
 | | The crystal structure of Rv2747 from Mycobacterium tuberculosis in complex with Acetyl CoA and L-Arginine | | Descriptor: | ACETYL COENZYME *A, ARGININE, Amino-acid acetyltransferase | | Authors: | Singh, E, Tiruttani Subhramanyam, U.K, Pal, R.K, Srinivasan, A, Gourinath, S, Das, U. | | Deposit date: | 2017-10-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural insights into the substrate binding mechanism of novel ArgA from Mycobacterium tuberculosis
Int. J. Biol. Macromol., 125, 2019
|
|
1CT5
 
 | | CRYSTAL STRUCTURE OF YEAST HYPOTHETICAL PROTEIN YBL036C-SELENOMET CRYSTAL | | Descriptor: | PROTEIN (YEAST HYPOTHETICAL PROTEIN, SELENOMET), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-08-18 | | Release date: | 1999-09-02 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a yeast hypothetical protein selected by a structural genomics approach.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3UP8
 
 | | Crystal structure of a putative 2,5-diketo-D-gluconic acid reductase B | | Descriptor: | ACETATE ION, Putative 2,5-diketo-D-gluconic acid reductase B | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a putative 2,5-diketo-D-gluconic acid reductase B
To be Published
|
|
3ULG
 
 | |
1F1M
 
 | | CRYSTAL STRUCTURE OF OUTER SURFACE PROTEIN C (OSPC) | | Descriptor: | OUTER SURFACE PROTEIN C, ZINC ION | | Authors: | Kumaran, D, Eswaramoorthy, S, Dunn, J.J, Swaminathan, S. | | Deposit date: | 2000-05-19 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of outer surface protein C (OspC) from the Lyme disease spirochete, Borrelia burgdorferi.
EMBO J., 20, 2001
|
|
4JOT
 
 | | Crystal structure of enoyl-CoA hydrotase from Deinococcus radiodurans R1 | | Descriptor: | Enoyl-CoA hydratase, putative, GLYCEROL | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-18 | | Release date: | 2013-04-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of enoyl-CoA hydrotase from Deinococcus radiodurans R1
To be Published
|
|
4K2N
 
 | | Crystal structure of an enoyl-CoA hydratase/ carnithine racemase from Magnetospirillum magneticum | | Descriptor: | Enoyl-CoA hydratase/carnithine racemase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-09 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase/ carnithine racemase from Magnetospirillum magneticum
To be Published
|
|
4HKE
 
 | | Crystal Structure of MoxT of Bacillus anthracis | | Descriptor: | Addiction module toxin component PemK, SULFATE ION | | Authors: | Verma, S, Kumar, S, Gourinath, S, Bhatnagar, R. | | Deposit date: | 2012-10-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of Bacillus anthracis MoxXT disruption and the modulation of MoxT ribonuclease activity by rationally designed peptides.
J.Biomol.Struct.Dyn., 33, 2015
|
|
5YB0
 
 | |
4HL9
 
 | | Crystal structure of antibiotic biosynthesis monooxygenase | | Descriptor: | Antibiotic biosynthesis monooxygenase | | Authors: | Rice, S, Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-10-16 | | Release date: | 2012-10-31 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of antibiotic biosynthesis monooxygenase
To be Published
|
|
5YD2
 
 | |
4HUJ
 
 | | Crystal structure of a hypothetical protein SMa0349 from Sinorhizobium meliloti | | Descriptor: | Uncharacterized protein | | Authors: | Rice, S, Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a hypothetical protein SMa0349 from Sinorhizobium meliloti
To be Published
|
|
1S5G
 
 | | Structure of Scallop myosin S1 reveals a novel nucleotide conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Risal, D, Gourinath, S, Himmel, D.M, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2004-01-20 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
5YII
 
 | |
4JCS
 
 | | Crystal structure of Enoyl-CoA hydratase/isomerase from Cupriavidus metallidurans CH34 | | Descriptor: | Enoyl-CoA hydratase/isomerase | | Authors: | Eswaramoorthy, S, Chamala, S, Chamala, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-22 | | Release date: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Enoyl-CoA hydratase/isomerase from Cupriavidus metallidurans CH34
To be Published
|
|
1SR6
 
 | | Structure of nucleotide-free scallop myosin S1 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Myosin essential light chain, ... | | Authors: | Risal, D, Gourinath, S, Himmel, D.M, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2004-03-22 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
4K29
 
 | | Crystal structure of an enoyl-CoA hydratase/isomerase from Xanthobacter autotrophicus Py2 | | Descriptor: | Enoyl-CoA hydratase/isomerase, GLYCEROL, L(+)-TARTARIC ACID | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-08 | | Release date: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase/isomerase from Xanthobacter autotrophicus Py2
TO BE PUBLISHED
|
|
