7GSY
 
 | |
7GT5
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000529a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, methyl [(3R,4S)-3-ethyl-4-hydroxy-1,1-dioxo-3,4-dihydro-1lambda~6~,2-benzothiazin-2(1H)-yl]acetate | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTS
 
 | |
7GSR
 
 | |
7GS7
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000621a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(1,2,3-thiadiazol-4-yl)phenyl ethylcarbamate, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTT
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000148a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-(3,4-dihydroquinoline-1(2H)-carbothioyl)propanamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GSA
 
 | |
7GSB
 
 | |
8U1E
 
 | |
9CYO
 
 | | Crystal structure of wild-type human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9CYQ
 
 | | Crystal structure of Q78R mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9CYR
 
 | | Crystal structure of D245G mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
9CYP
 
 | | Crystal structure of I19V mutant human PTP1B (PTPN1) at room temperature (298 K) | | Descriptor: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Ebrahim, A, Perdikari, A, Woods, V.A, Lawler, K, Bounds, R, Singh, N.I, Mehlman, T, Riley, B.T, Sharma, S, Morris, J.W, Keogh, J.M, Henning, E, Smith, M, Farooqi, I.S, Keedy, D.A. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structures of human PTP1B variants reveal allosteric sites to target for weight loss therapy.
Biorxiv, 2024
|
|
6QJZ
 
 | |
3NVJ
 
 | | Crystal structure of the C143A/C166A mutant of Ero1p | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, CADMIUM ION, Endoplasmic oxidoreductin-1, ... | | Authors: | Fass, D, Vonshak, O. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Steps in reductive activation of the disulfide-generating enzyme Ero1p
Protein Sci., 19, 2010
|
|
7R65
 
 | |
6YBH
 
 | | Deoxyribonucleoside Kinase | | Descriptor: | Deoxynucleoside kinase, GLYCEROL, Pyrrolo-dC, ... | | Authors: | Allouche-Arnon, H, Bar-Shir, A, Dym, O. | | Deposit date: | 2020-03-17 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computationally designed dual-color MRI reporters for noninvasive imaging of transgene expression.
Nat.Biotechnol., 40, 2022
|
|
6ZBS
 
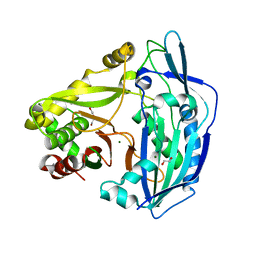 | | Beta ODAP Synthetase (BOS) | | Descriptor: | Beta ODAP Synthetase (BOS), DI(HYDROXYETHYL)ETHER, MAGNESIUM ION | | Authors: | Dym, O. | | Deposit date: | 2020-06-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification and characterization of the key enzyme in the biosynthesis of the neurotoxin beta-ODAP in grass pea.
J.Biol.Chem., 298, 2022
|
|
8FNW
 
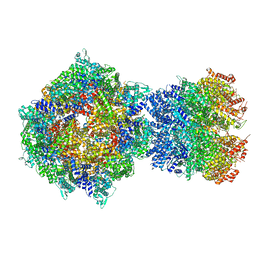 | | Structure of RdrA-RdrB complex from Escherichia coli RADAR defense system | | Descriptor: | Adenosine deaminase, Archaeal ATPase, ZINC ION | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNT
 
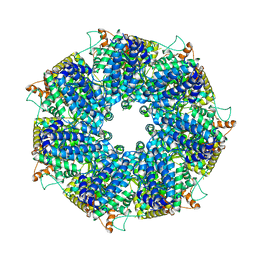 | | Structure of RdrA from Escherichia coli RADAR defense system | | Descriptor: | Archaeal ATPase | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNV
 
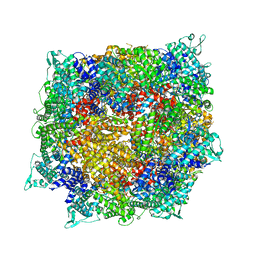 | | Structure of RdrB from Escherichia coli RADAR defense system | | Descriptor: | Adenosine deaminase, ZINC ION | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
8FNU
 
 | | Structure of RdrA from Streptococcus suis RADAR defense system | | Descriptor: | KAP NTPase domain-containing protein | | Authors: | Duncan-Lowey, B, Johnson, A.G, Rawson, S, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2022-12-28 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of the RADAR supramolecular anti-phage defense complex.
Cell, 186, 2023
|
|
3M31
 
 | |
3T58
 
 | |
3T59
 
 | |
