3BCK
 
 | |
3HCF
 
 | |
3HCE
 
 | | Crystal Structure of E185D hPNMT in Complex With Octopamine and AdoHcy | | Descriptor: | 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
5DCH
 
 | | Crystal structure of Pseudomonas aeruginosa DsbA E82I in complex with MIPS-0000851 (3-[(2-METHYLBENZYL)SULFANYL]-4H-1,2,4-TRIAZOL-4-AMINE) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, GLYCEROL, ... | | Authors: | McMahon, R.M, Martin, J.L. | | Deposit date: | 2015-08-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
3HCB
 
 | | Crystal Structure of hPNMT in Complex With Noradrenochrome and AdoHcy | | Descriptor: | (3S)-3-hydroxy-2,3-dihydro-1H-indole-5,6-dione, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L, Gee, C.L. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
3HCD
 
 | | Crystal Structure of hPNMT in Complex With Noradrenaline and AdoHcy | | Descriptor: | L-NOREPINEPHRINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
3HCA
 
 | | Crystal Structure of E185Q hPNMT in Complex With Octopamine and AdoHcy | | Descriptor: | 1,2-ETHANEDIOL, 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, Phenylethanolamine N-methyltransferase, ... | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
3HCC
 
 | | Crystal Structure of hPNMT in Complex With anti-9-amino-5-(trifluromethyl) benzonorbornene and AdoHcy | | Descriptor: | (1S,4R,9S)-5-(trifluoromethyl)-1,2,3,4-tetrahydro-1,4-methanonaphthalen-9-amine, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L, Gee, C.L, Puri, M. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
1N7I
 
 | | The structure of Phenylethanolamine N-methyltransferase in complex with S-adenosylhomocysteine and the inhibitor LY134046 | | Descriptor: | 8,9-DICHLORO-2,3,4,5-TETRAHYDRO-1H-BENZO[C]AZEPINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMillan, F.M, Archbold, J, McLeish, M.J, Caine, J.M, Criscione, K.R, Grunewald, G.L, Martin, J.L. | | Deposit date: | 2002-11-15 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular recognition of sub-micromolar inhibitors by the epinephrine-synthesizing enzyme phenylethanolamine N-methyltransferase.
J.Med.Chem., 47, 2004
|
|
1N7J
 
 | | The structure of Phenylethanolamine N-methyltransferase in complex with S-adenosylhomocysteine and an iodinated inhibitor | | Descriptor: | 7-IODO-1,2,3,4-TETRAHYDRO-ISOQUINOLINE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMillan, F.M, Archbold, J, McLeish, M.J, Caine, J.M, Criscione, K.R, Grunewald, G.L, Martin, J.L. | | Deposit date: | 2002-11-15 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular recognition of sub-micromolar inhibitors by the epinephrine-synthesizing enzyme phenylethanolamine N-methyltransferase.
J.Med.Chem., 47, 2004
|
|
6C29
 
 | | Crystal structure of the N-terminal periplasmic domain of ScsB from Proteus mirabilis | | Descriptor: | Putative metal resistance protein | | Authors: | Furlong, E.J, Choudhury, H.G, Kurth, F, Martin, J.L. | | Deposit date: | 2018-01-07 | | Release date: | 2018-03-07 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | Disulfide isomerase activity of the dynamic, trimericProteus mirabilisScsC protein is primed by the tandem immunoglobulin-fold domain of ScsB.
J. Biol. Chem., 293, 2018
|
|
1MTR
 
 | | HIV-1 PROTEASE COMPLEXED WITH A CYCLIC PHE-ILE-VAL PEPTIDOMIMETIC INHIBITOR | | Descriptor: | HIV-1 PROTEASE, SULFATE ION, [1-BENZYL-3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2] HEPTADECA-1(16),13(17),14-TRIEN-11-YLAMINO)-2-HYDROXY-PROPYL]-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Wickramasinghe, W, Begun, J, Martin, J.L. | | Deposit date: | 1996-02-15 | | Release date: | 1996-08-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate-based cyclic peptidomimetics of Phe-Ile-Val that inhibit HIV-1 protease using a novel enzyme-binding mode.
J.Am.Chem.Soc., 118, 1996
|
|
1PEN
 
 | | ALPHA-CONOTOXIN PNI1 | | Descriptor: | ALPHA-CONOTOXIN PNIA | | Authors: | Hu, S.-H, Gehrmann, J, Guddat, L.W, Alewood, P.F, Craik, D.J, Martin, J.L. | | Deposit date: | 1996-01-29 | | Release date: | 1997-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A crystal structure of the neuronal acetylcholine receptor antagonist, alpha-conotoxin PnIA from Conus pennaceus.
Structure, 4, 1996
|
|
1NOT
 
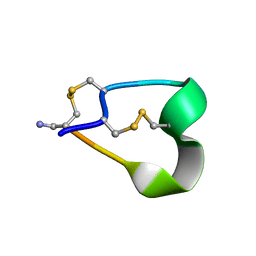 | | THE 1.2 ANGSTROM STRUCTURE OF G1 ALPHA CONOTOXIN | | Descriptor: | GI ALPHA CONOTOXIN | | Authors: | Guddat, L.W, Shan, L, Martin, J.L, Edmundson, A.B, Gray, W.R. | | Deposit date: | 1996-05-02 | | Release date: | 1996-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Three-dimensional structure of the alpha-conotoxin GI at 1.2 A resolution
Biochemistry, 35, 1996
|
|
4GUX
 
 | | Crystal structure of trypsin:MCoTi-II complex | | Descriptor: | ACETATE ION, CALCIUM ION, Cationic trypsin, ... | | Authors: | King, G.J, Daly, N.L, Thorstholm, L, Greenwood, K.P, Rosengren, K.J, Heras, B, Craik, D.J, Martin, J.L. | | Deposit date: | 2012-08-30 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural insights into the role of the cyclic backbone in a squash trypsin inhibitor
J.Biol.Chem., 288, 2013
|
|
1UTE
 
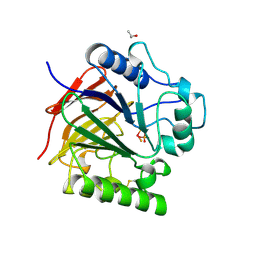 | | PIG PURPLE ACID PHOSPHATASE COMPLEXED WITH PHOSPHATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ISOPROPYL ALCOHOL, MU-OXO-DIIRON, ... | | Authors: | Guddat, L.W, Mcalpine, A, Hume, D, Hamilton, S, De Jersey, J, Martin, J.L. | | Deposit date: | 1999-01-18 | | Release date: | 1999-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of mammalian purple acid phosphatase.
Structure Fold.Des., 7, 1999
|
|
4DVC
 
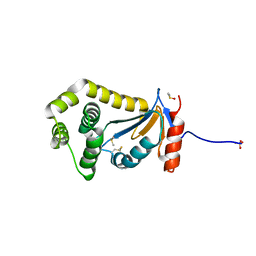 | | Structural and functional studies of TcpG, the Vibrio cholerae DsbA disulfide-forming protein required for pilus and cholera toxin production | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Thiol:disulfide interchange protein DsbA | | Authors: | Walden, P.M, Martin, J.L. | | Deposit date: | 2012-02-23 | | Release date: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A resolution crystal structure of TcpG, the Vibrio cholerae DsbA disulfide-forming protein required for pilus and cholera-toxin production
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4I1K
 
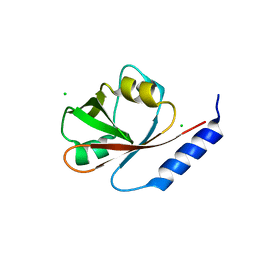 | | Crystal Structure of VRN1 (Residues 208-341) | | Descriptor: | B3 domain-containing transcription factor VRN1, CHLORIDE ION | | Authors: | King, G, Chanson, A.H, McCallum, E.J, Ohme-Takagi, M, Byriel, K, Hill, J.M, Martin, J.L, Mylne, J.S. | | Deposit date: | 2012-11-21 | | Release date: | 2012-12-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Arabidopsis B3 Domain Protein VERNALIZATION1 (VRN1) Is Involved in Processes Essential for Development, with Structural and Mutational Studies Revealing Its DNA-binding Surface.
J.Biol.Chem., 288, 2013
|
|
3PUJ
 
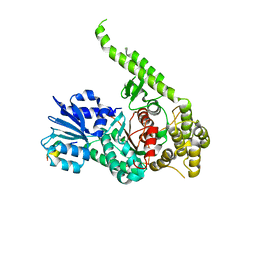 | | Crystal structure of the MUNC18-1 and SYNTAXIN4 N-Peptide complex | | Descriptor: | Syntaxin-4 N-terminal peptide, Syntaxin-binding protein 1 | | Authors: | Hu, S.-H, Christie, M.P, Saez, N.J, Latham, C.F, Jarrott, R, Lua, L.H.L, Collins, B.M, Martin, J.L. | | Deposit date: | 2010-12-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.313 Å) | | Cite: | Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7RGV
 
 | |
4OCF
 
 | |
4OCE
 
 | |
4P3Y
 
 | |
4OD7
 
 | |
1G9T
 
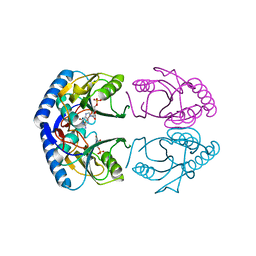 | | CRYSTAL STRUCTURE OF E.COLI HPRT-GMP COMPLEX | | Descriptor: | ANY 5'-MONOPHOSPHATE NUCLEOTIDE, GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Guddat, L.W, Vos, S, Martin, J.L, keough, D.T, de Jersey, J. | | Deposit date: | 2000-11-28 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of free, IMP-, and GMP-bound Escherichia coli hypoxanthine phosphoribosyltransferase.
Protein Sci., 11, 2002
|
|
