8WTI
 
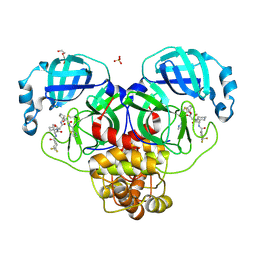 | | Crystal structure of the SARS-CoV-2 main protease in complex with 20j | | Descriptor: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Zeng, R, Zhao, X, Yang, S.Y, Lei, J. | | Deposit date: | 2023-10-18 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of alpha-Ketoamide inhibitors of SARS-CoV-2 main protease derived from quaternized P1 groups.
Bioorg.Chem., 143, 2024
|
|
8JOP
 
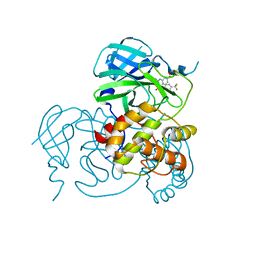 | | Crystal structure of the SARS-CoV-2 main protease in complex with 11a | | Descriptor: | 3C-like proteinase nsp5, methyl (6~{R})-5-ethanoyl-7-oxidanylidene-6-[4-(trifluoromethyl)phenyl]-8,9,10,11-tetrahydro-6~{H}-benzo[b][1,4]benzodiazepine-2-carboxylate | | Authors: | Zeng, R, Liu, Y.Z, Wang, F.L, Yang, S.Y, Lei, J. | | Deposit date: | 2023-06-08 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of benzodiazepine derivatives as a new class of covalent inhibitors of SARS-CoV-2 main protease.
Bioorg.Med.Chem.Lett., 92, 2023
|
|
8XY4
 
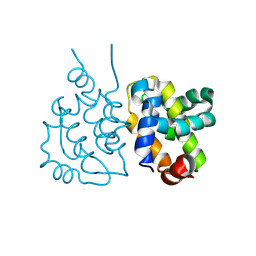 | | Crystal structure of VACV N1 protein | | Descriptor: | N1L | | Authors: | Ni, X.C, Lei, J. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into MPXV P1 protein and its orthologs reveal conformational dynamics and a conserved antiviral pocket.
Emerg Microbes Infect, 14, 2025
|
|
8XY1
 
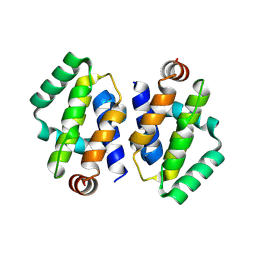 | | Crystal structure of MPXV P1 protein | | Descriptor: | Protein OPG035 | | Authors: | Ni, X.C, Lei, J. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.575 Å) | | Cite: | Structural insights into MPXV P1 protein and its orthologs reveal conformational dynamics and a conserved antiviral pocket.
Emerg Microbes Infect, 14, 2025
|
|
8XY3
 
 | | Crystal structure of VARV P1 protein | | Descriptor: | P1L | | Authors: | Ni, X.C, Lei, J. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into MPXV P1 protein and its orthologs reveal conformational dynamics and a conserved antiviral pocket.
Emerg Microbes Infect, 14, 2025
|
|
8Y7T
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with C2 | | Descriptor: | 3C-like proteinase nsp5, 6-(iminomethyl)-4-(2-pyridin-2-ylethyl)-2-[4-(trifluoromethyl)phenyl]-1,2,4-triazine-3,5-dione | | Authors: | Zeng, R, Deng, X.Y, Yang, Z.Y, Wang, K, Jiang, Y.Y, Lei, J. | | Deposit date: | 2024-02-05 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A deep learning model for structure-based bioactivity optimization and its application in the bioactivity optimization of a SARS-CoV-2 main protease inhibitor.
Eur.J.Med.Chem., 291, 2025
|
|
8XY2
 
 | | Crystal structure of MPXV P1 protein S87P mutant | | Descriptor: | Protein OPG035 | | Authors: | Ni, X.C, Lei, J. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural insights into MPXV P1 protein and its orthologs reveal conformational dynamics and a conserved antiviral pocket.
Emerg Microbes Infect, 14, 2025
|
|
8Y7U
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with C5 | | Descriptor: | 2-(3-fluoro-4-(trifluoromethyl)phenyl)-6-(iminomethyl)-4-(2-oxo-2-(pyridin-2-yl)ethyl)-1,2,4-triazine-3,5(2H,4H)-dione, 3C-like proteinase nsp5 | | Authors: | Zeng, R, Deng, X.Y, Yang, Z.Y, Wang, K, Jiang, Y.Y, Lei, J. | | Deposit date: | 2024-02-05 | | Release date: | 2025-02-05 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A deep learning model for structure-based bioactivity optimization and its application in the bioactivity optimization of a SARS-CoV-2 main protease inhibitor.
Eur.J.Med.Chem., 291, 2025
|
|
8ILC
 
 | | Crystal structure of Se-Met CoV-Y domain of Nsp3 in SARS-CoV-2 | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION | | Authors: | Wang, K, Nan, J, Lei, J. | | Deposit date: | 2023-03-03 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The CoV-Y domain of SARS-CoV-2 Nsp3 interacts with BRAP to stimulate NF-kappa B signaling and induce host inflammatory responses.
Int.J.Biol.Macromol., 280, 2024
|
|
8I30
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with 32j | | Descriptor: | (2~{R})-1-[4,4-bis(fluoranyl)cyclohexyl]carbonyl-4,4-bis(fluoranyl)-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]pyrrolidine-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Zeng, R, Huang, C, Xie, L.W, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | Deposit date: | 2023-01-16 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and structure-activity relationship studies of novel alpha-ketoamide derivatives targeting the SARS-CoV-2 main protease.
Eur.J.Med.Chem., 259, 2023
|
|
8X54
 
 | | Cryo-EM structure of human gamma-secretase in complex with APP-C99 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-11-16 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism of substrate recognition and cleavage by human gamma-secretase.
Science, 384, 2024
|
|
8X52
 
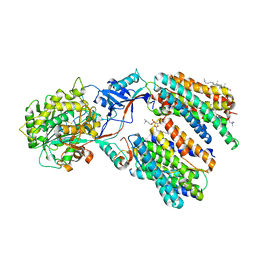 | | Cryo-EM structure of human gamma-secretase in complex with Abeta49 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-11-16 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism of substrate recognition and cleavage by human gamma-secretase.
Science, 384, 2024
|
|
8X53
 
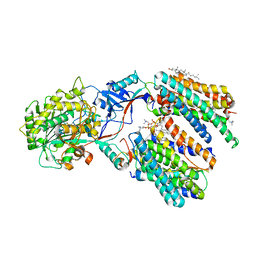 | | Cryo-EM structure of human gamma-secretase in complex with Abeta46 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2023-11-16 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular mechanism of substrate recognition and cleavage by human gamma-secretase.
Science, 384, 2024
|
|
3J2G
 
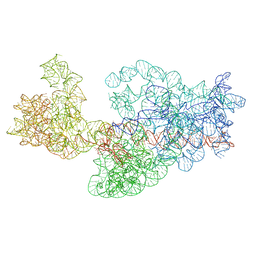 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (16.5 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process.
Nucleic Acids Res., 41, 2013
|
|
3J2E
 
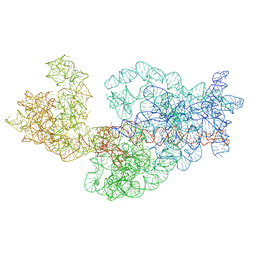 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (15.3 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J2D
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (18.700001 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J28
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (12.9 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J2C
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA body domain, 16S rRNA head domain | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (13.2 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J29
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J2F
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (17.6 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process.
Nucleic Acids Res., 41, 2013
|
|
7Y7Y
 
 | | Cryo-EM structure of human GABA transporter GAT1 bound with nipecotic acid in NaCl solution in an inward-occluded state at 2.4 angstrom | | Descriptor: | (3R)-piperidine-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhu, A, Huang, J, Kong, F, Tan, J, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-06-22 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Molecular basis for substrate recognition and transport of human GABA transporter GAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Y7W
 
 | | Cryo-EM structure of human GABA transporter GAT1 bound with GABA in NaCl solution in an inward-occluded state at 2.4 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Zhu, A, Huang, J, Kong, F, Tan, J, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-06-22 | | Release date: | 2023-04-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Molecular basis for substrate recognition and transport of human GABA transporter GAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Y7V
 
 | | Cryo-EM structure of human apo GABA transporter GAT1 in an inward-open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Sodium- and chloride-dependent GABA transporter 1 | | Authors: | Zhu, A, Huang, J, Kong, F, Tan, J, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-06-22 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Molecular basis for substrate recognition and transport of human GABA transporter GAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Y7Z
 
 | | Cryo-EM structure of human GABA transporter GAT1 bound with tiagabine in NaCl solution in an inward-open state at 3.2 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Sodium- and chloride-dependent GABA transporter 1, ... | | Authors: | Zhu, A, Huang, J, Kong, F, Tan, J, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2022-06-22 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis for substrate recognition and transport of human GABA transporter GAT1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Y5X
 
 | | CryoEM structure of PS2-containing gamma-secretase treated with MRK-560 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-11-02 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
