8Y8X
 
 | | Cryo-EM structure of GlpF with P177 deletion in POPC nanodisc | | Descriptor: | Glycerol uptake facilitator protein | | Authors: | Kozai, D, Suzuki, S, Kamegawa, A, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Narrowed pore conformations of aquaglyceroporins AQP3 and GlpF.
Nat Commun, 16, 2025
|
|
8Y8S
 
 | | Cryo-EM structure of AQP3 in DMPC nanodisc | | Descriptor: | Aquaporin-3 | | Authors: | Kozai, D, Suzuki, S, Kamegawa, A, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Narrowed pore conformations of aquaglyceroporins AQP3 and GlpF.
Nat Commun, 16, 2025
|
|
8Y8R
 
 | | Cryo-EM structure of AQP3 Y212T in DDM micelle | | Descriptor: | Aquaporin-3, DODECYL-BETA-D-MALTOSIDE | | Authors: | Kozai, D, Suzuki, S, Kamegawa, A, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Narrowed pore conformations of aquaglyceroporins AQP3 and GlpF.
Nat Commun, 16, 2025
|
|
8Y8P
 
 | | Cryo-EM structure of AQP3 Y212F in POPC nanodisc | | Descriptor: | Aquaporin-3 | | Authors: | Kozai, D, Suzuki, S, Kamegawa, A, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Narrowed pore conformations of aquaglyceroporins AQP3 and GlpF.
Nat Commun, 16, 2025
|
|
8Y8Q
 
 | | Cryo-EM structure of AQP3 Y212T in POPC nanodisc | | Descriptor: | Aquaporin-3 | | Authors: | Kozai, D, Suzuki, S, Kamegawa, A, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Narrowed pore conformations of aquaglyceroporins AQP3 and GlpF.
Nat Commun, 16, 2025
|
|
8Y8O
 
 | | Cryo-EM structure of AQP3 in POPC nanodisc | | Descriptor: | Aquaporin-3 | | Authors: | Kozai, D, Suzuki, S, Kamegawa, A, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Narrowed pore conformations of aquaglyceroporins AQP3 and GlpF.
Nat Commun, 16, 2025
|
|
8Y8N
 
 | | Cryo-EM structure of AQP3 in DDM micelle | | Descriptor: | Aquaporin-3 | | Authors: | Kozai, D, Suzuki, S, Kamegawa, A, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2025-03-19 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Narrowed pore conformations of aquaglyceroporins AQP3 and GlpF.
Nat Commun, 16, 2025
|
|
8Y8W
 
 | | Cryo-EM structure of GlpF in POPC nanodisc | | Descriptor: | Glycerol uptake facilitator protein | | Authors: | Kozai, D, Suzuki, S, Kamegawa, A, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2025-03-19 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Narrowed pore conformations of aquaglyceroporins AQP3 and GlpF.
Nat Commun, 16, 2025
|
|
8Y8V
 
 | | Cryo-EM structure of AQP7 in POPC nanodisc | | Descriptor: | Green fluorescent protein,Aquaporin-7, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine | | Authors: | Kozai, D, Suzuki, S, Kamegawa, A, Nishikawa, K, Suzuki, H, Fujiyosh, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2025-03-19 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Narrowed pore conformations of aquaglyceroporins AQP3 and GlpF.
Nat Commun, 16, 2025
|
|
5Z7Y
 
 | | Crystal structure of Striga hermonthica HTL7 (ShHTL7) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, Hyposensitive to light 7, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
9IZF
 
 | | Cryo-EM structure of LPA1-Gi complex with LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Suzuki, S, Nishikawa, K, Kamegawa, A, Hiroaki, Y, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2024-08-01 | | Release date: | 2025-01-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural insights into the engagement of lysophosphatidic acid receptor 1 with different G proteins.
J.Struct.Biol., 217, 2024
|
|
9IZH
 
 | | Cryo-EM structure of LPA1-G13 complex with LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, G protein subunit 13 (Gi2-mini-G13 chimera), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Suzuki, S, Nishikawa, K, Kamegawa, A, Hiroaki, Y, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2024-08-01 | | Release date: | 2025-01-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural insights into the engagement of lysophosphatidic acid receptor 1 with different G proteins.
J.Struct.Biol., 217, 2024
|
|
5Z7X
 
 | | Crystal structure of Striga hermonthica HTL4 (ShHTL4) | | Descriptor: | 1,2-ETHANEDIOL, Hyposensitive to light 4, MAGNESIUM ION | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
8ZVO
 
 | | AtKAI2 apo structure | | Descriptor: | GLYCEROL, Probable esterase KAI2, SULFATE ION | | Authors: | Takegamie, K, Takeuchi, J, Nakamura, A. | | Deposit date: | 2024-06-11 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural requirements of KAI2 ligands for activation of signal transduction.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8ZVN
 
 | | AtKAI2 (+)-6'-carba-dMGer complex | | Descriptor: | (2~{S})-2-[(2-nitro-4-phenyl-phenyl)methyl]-2~{H}-furan-5-one, GLYCEROL, Probable esterase KAI2, ... | | Authors: | Takegamie, K, Kushihara, R, Takeuchi, J, Nakamura, A. | | Deposit date: | 2024-06-11 | | Release date: | 2025-02-19 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural requirements of KAI2 ligands for activation of signal transduction.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
5Z7Z
 
 | | Crystal structure of Striga hermonthica Dwarf14 (ShD14) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dwarf 14, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
5Z7W
 
 | | Crystal structure of Striga hermonthica HTL1 (ShHTL1) | | Descriptor: | GLYCEROL, Hyposensitive to light 1, MAGNESIUM ION, ... | | Authors: | Xu, Y, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga.
Nat Commun, 9, 2018
|
|
8JT7
 
 | | Structure of arginine oxidase from Pseudomonas sp. TRU 7192 | | Descriptor: | Amine oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yamaguchi, H, Numoto, N, Suzuki, H, Nishikawa, K, Kamegawa, A, Takahashi, K, Sugiki, M, Fujiyoshi, Y. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-26 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Open and closed structures of L-arginine oxidase by cryo-electron microscopy and X-ray crystallography.
J.Biochem., 177, 2025
|
|
5YAQ
 
 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity complexed with scyllo-inosose | | Descriptor: | (2R,3S,4s,5R,6S)-2,3,4,5,6-pentahydroxycyclohexanone, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2017-09-01 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
5YAB
 
 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Descriptor: | ACETATE ION, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2017-08-31 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
5YAP
 
 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity complexed with L-glucono-1,5-lactone | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-glucono-1,5-lactone, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2017-09-01 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
5YA8
 
 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity complexed with myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2017-08-31 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
4WJ4
 
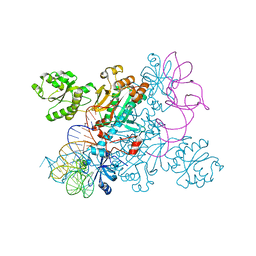 | | Crystal structure of non-discriminating aspartyl-tRNA synthetase from Pseudomonas aeruginosa complexed with tRNA(Asn) and aspartic acid | | Descriptor: | 76mer-tRNA, ASPARTIC ACID, Aspartate--tRNA(Asp/Asn) ligase | | Authors: | Suzuki, T, Nakamura, A, Kato, K, Tanaka, I, Yao, M. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structure of the Pseudomonas aeruginosa transamidosome reveals unique aspects of bacterial tRNA-dependent asparagine biosynthesis
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WFT
 
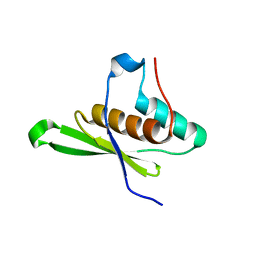 | | Crystal structure of tRNA-dihydrouridine(20) synthase dsRBD domain | | Descriptor: | tRNA-dihydrouridine(20) synthase [NAD(P)+]-like | | Authors: | Bou-Nader, C, Pecqueur, L, Kamah, A, Bregeon, D, Golinelli-Pimpaneau, B, Guimaraes, B.G, Fontecave, M, Hamdane, D. | | Deposit date: | 2014-09-17 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An extended dsRBD is required for post-transcriptional modification in human tRNAs.
Nucleic Acids Res., 43, 2015
|
|
2Z0N
 
 | | Crystal structure of APPL1-BAR domain | | Descriptor: | DCC-interacting protein 13-alpha | | Authors: | Murayama, K, Kato-Murayama, M, Sakamoto, A, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of APPL1-BAR domain
To be Published
|
|
