2KCT
 
 | | Solution nmr structure of the ob-fold domain of heme chaperone ccme from desulfovibrio vulgaris. northeast structural genomics target dvr115g. | | Descriptor: | Cytochrome c-type biogenesis protein CcmE | | Authors: | Aramini, J.M, Rossi, P, Lee, H, Lemak, A, Wang, H, Foote, E.L, Jiang, M, Xiao, R, Nair, R, Swapna, G.V.T, Acton, T.B, Rost, B, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution nmr structure of the ob-fold domain of heme chaperone
ccme from desulfovibrio vulgaris. northeast structural genomics target dvr115g.
To be Published
|
|
2K5N
 
 | | Solution NMR Structure of the N-Terminal Domain of Protein ECA1580 from Erwinia carotovora, Northeast Structural Genomics Consortium Target EwR156A | | Descriptor: | Putative cold-shock protein | | Authors: | Mills, J.L, Eletsky, A, Zhang, Q, Lee, D, Jiang, M, Ciccosanti, C, Xiao, R, Lui, J, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Putative Cold Shock Protein from Erwinia carotovora: Northeast Structural Genomics Consortium Target EwR156a
To be Published
|
|
2KK8
 
 | | NMR Solution Structure of a Putative Uncharacterized Protein obtained from Arabidopsis thaliana: Northeast Structural Genomics Consortium target AR3449A | | Descriptor: | Uncharacterized protein AT4g05270 | | Authors: | Mani, R, Gurla, S.V.T, Shastry, R, Ciccosanti, C, Foote, E, Jiang, M, Xiao, R, Nair, R, Everett, J, Huang, Y, Acton, T, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-16 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a Putative Uncharacterized Protein obtained from Arabidopsis thaliana: Northeast Structural Genomics Consortium Target AR3449A
To be Published
|
|
2K5D
 
 | | SOLUTION NMR STRUCTURE OF SAG0934 from Streptococcus agalactiae. NORTHEAST STRUCTURAL GENOMICS TARGET SaR32[1-108]. | | Descriptor: | uncharacterized protein SAG0934 | | Authors: | Aramini, J.M, Rossi, P, Zhao, L, Foote, E.L, Jiang, M, Xiao, R, Sharma, S, Swapna, G.VT, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-26 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION NMR STRUCTURE OF SAG0934 from Streptococcus agalactiae. NORTHEAST STRUCTURAL GENOMICS TARGET SaR32[1-108].
To be Published
|
|
2KJR
 
 | | Solution NMR structure of the N-terminal Ubiquitin-like Domain from Tubulin-binding Cofactor B, CG11242, from Drosophila melanogaster. Northeast Structural Genomics Consortium Target FR629A (residues 8-92) | | Descriptor: | CG11242 | | Authors: | Ramelot, T.A, Cort, J.R, Shastry, R, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Swapna, G, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-08 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the N-terminal Ubiquitin-like Domain from
Tubulin-binding Cofactor B, CG11242, from Drosophila melanogaster. Northeast
Structural Genomics Consortium Target FR629A (residues 8-92)
To be Published
|
|
2KHV
 
 | | Solution NMR structure of protein Nmul_A0922 from Nitrosospira multiformis. Northeast Structural Genomics Consortium target NmR38B. | | Descriptor: | Phage integrase | | Authors: | Wu, Y, Mills, J.L, Wang, D, Ciccosanti, C, Sukumaran, D.K, Jiang, M, Garcia, E, Nair, R, Rost, B, Swapna, G.V.T, Acton, T.B, Xiao, R, Everett, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-13 | | Release date: | 2009-04-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein Nmul_A0922 from Nitrosospira multiformis. Northeast Structural Genomics Consortium target NmR38B.
To be Published
|
|
2KJ6
 
 | | NMR Solution Structure of a Tubulin folding cofactor B obtained from Arabidopsis thaliana: Northeast Structural Genomics Consortium target AR3436A | | Descriptor: | Tubulin folding cofactor B | | Authors: | Mani, R, Swapna, G.V.T, Shastry, R, Foote, E, Ciccosanti, C, Jiang, M, Xiao, R, Nair, R, Everett, J, Huang, Y.J, Acton, T, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-05-22 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Tbulin folding Cofactor B obtained from Arabidopsis thaliana: Northeast
To be Published
|
|
2KC7
 
 | | Solution NMR structure of Bacteroides fragilis protein BF1650. Northeast Structural Genomics Consortium target BfR218 | | Descriptor: | bfr218_protein | | Authors: | Eletsky, A, Wu, Y, Sukumaran, D, Lee, H, Lee, D.Y, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-17 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Bacteroides fragilis protein BF1650. Northeast Structural Genomics Consortium target BfR218
To be Published
|
|
2KD7
 
 | | Solution NMR structure of F5/8 type C-terminal domain of a putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium target BtR324B | | Descriptor: | Putative chitobiase | | Authors: | Eletsky, A, Mills, J.L, Lee, H, Lee, D, Jiang, M, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-01-04 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of F5/8 type C-terminal domain of a putative chitobiase from Bacteroides thetaiotaomicron.
To be Published
|
|
2KLB
 
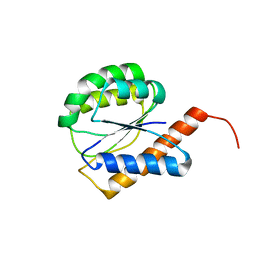 | | NMR Solution structure of a diflavin flavoprotein A3 from Nostoc sp. PCC 7120, Northeast Structural Genomics Consortium Target NsR431C | | Descriptor: | Putative diflavin flavoprotein A 3 | | Authors: | Swapna, G.V.T, Ciccosanti, C, Wang, D, Jiang, M, Xiao, R, Acton, T.B, Everett, J.K, Nair, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution structure of a diflavin flavoprotein A3 from Nostoc sp. PCC 7120, Northeast Structural Genomics Consortium Target NsR431C
To be Published
|
|
2K4Y
 
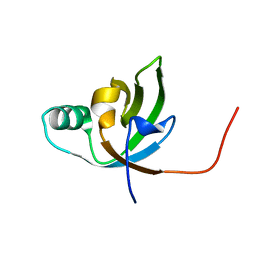 | | NMR Structure of FeoA-like protein from Clostridium acetobutylicum: Northeast Structural Genomics Consortium Target CaR178 | | Descriptor: | FeoA-like protein | | Authors: | Singarapu, K, Wu, Y, Hua, J, Sukumaran, D, Zhao, L, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Baran, M.C, Swapna, G.V.T, Acton, T, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of FeoA-like protein from Clostridium acetobutylicum: Northeast Structural Genomics Consortium Target CaR178
To be Published
|
|
2K5G
 
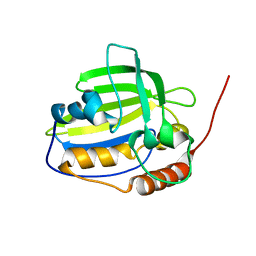 | | Solution NMR structure of protein encoded by gene BPP1335 from Bordetella parapertussis: Northeast Structural Genomics Target BpR195 | | Descriptor: | uncharacterized protein | | Authors: | Singarapu, K, Eletsky, A, Sathyamoorthy, B, Sukumaran, D, Wang, D, Jiang, M, Ciccosanti, C, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-27 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein encoded by gene BPP1335 from
Bordetella parapertussis: Northeast Structural Genomics Target BpR195
To be Published
|
|
2KKS
 
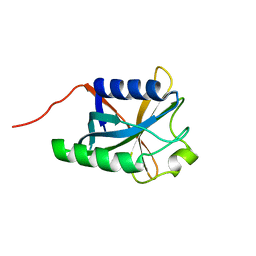 | | Solution Structure Of Protein DSY2949 From Desulfitobacterium hafniense. Northeast Structural Genomics Consortium Target DhR27 | | Descriptor: | Uncharacterized protein | | Authors: | Wu, Y, Mills, J.L, Wang, H, Ciccosanti, C, Jiang, M, Sukumaran, D, Zhang, Q, Nair, R, Rost, B, Acton, T, Xiao, R, Swapna, G.V.T, Everett, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of Protein DSY2949 From Desulfitobacterium hafniense. Northeast Structural Genomics Consortium Target DhR27
To be Published
|
|
2KEY
 
 | | Solution NMR structure of a domain from a putative phage integrase protein BF2284 from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR257C | | Descriptor: | Putative phage integrase | | Authors: | Mills, J.L, Sathyamoorthy, B, Sukumaran, D.K, Lee, D, Ciccosanti, C, Jiang, M, Xiao, R, Acton, T.B, Swapna, G.V.T, Rost, B, Nair, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-06 | | Release date: | 2009-04-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Domain from a Putative Phage Integrase Protein, BF2284, from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR257C
To be Published
|
|
6IQT
 
 | | Crystal Structure of CagV, a VirB8 homolog of T4SS from Helicobacter pylori Strain 26695 | | Descriptor: | Cag pathogenicity island protein (Cag10) | | Authors: | Wu, X, Zhao, Y, Sun, L, Jiang, M, Wang, Q, Wang, Q, Yang, W, Wu, Y. | | Deposit date: | 2018-11-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Crystal structure of CagV, the Helicobacter pylori homologue of the T4SS protein VirB8.
Febs J., 286, 2019
|
|
8I5D
 
 | | Crystal structure of a TCR in complex with HLA-A*11:01 bound to KRAS peptide (VVGAVGVGK) | | Descriptor: | Beta-2-microglobulin, MHC class I antigen (Fragment), TCR alpha chain, ... | | Authors: | Lu, D, Chen, Y, Jiang, M, Tan, S.G, Chai, Y, Gao, G.F. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of a TCR in complex with HLA-A*11:01 bound to KRAS peptide (VVGAVGVGK)
Nat Commun, 2023
|
|
8I5C
 
 | | Crystal structure of a TCR in complex with HLA-A*11:01 bound to KRAS peptide (VVGAVGVGK) | | Descriptor: | Beta-2-microglobulin, MHC class I antigen (Fragment), TCR alpha chain, ... | | Authors: | Lu, D, Chen, Y, Jiang, M, Tan, S.G, Chai, Y, Gao, G.F. | | Deposit date: | 2023-01-24 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Crystal structure of a TCR in complex with HLA-A*11:01 bound to KRAS peptide (VVGAVGVGK)
Nat Commun, 2023
|
|
8I5E
 
 | | Crystal structure of HLA-A*11:01 in complex with KRAS peptide (VVGAGGVGK) | | Descriptor: | Beta-2-microglobulin, KRAS-G12wt-9, MHC class I antigen (Fragment) | | Authors: | Lu, D, Chen, Y, Jiang, M, Tan, S.G, Chai, Y, Gao, G.F. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a TCR in complex with HLA-A*11:01 bound to KRAS peptide (VVGAVGVGK)
Nat Commun, 2023
|
|
5CFF
 
 | | Crystal structure of Miranda/Staufen dsRBD5 complex | | Descriptor: | Miranda, Staufen | | Authors: | Shan, Z, Wen, W. | | Deposit date: | 2015-07-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis of Miranda-mediated Staufen localization during Drosophila neuroblast asymmetric division
Nat Commun, 6, 2015
|
|
6VU2
 
 | | M1214_N1 Fab structure | | Descriptor: | M1214 N1 Fab heavy chain, M1214 N1 Fab light chain | | Authors: | Pan, R, Kong, X. | | Deposit date: | 2020-02-14 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | VSV-Displayed HIV-1 Envelope Identifies Broadly Neutralizing Antibodies Class-Switched to IgG and IgA.
Cell Host Microbe, 27, 2020
|
|
6VY2
 
 | |
8TNL
 
 | |
6J4R
 
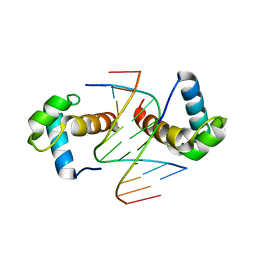 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*C)-3'), DNA (5'-D(*CP*CP*AP*TP*AP*TP*AP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
6J4K
 
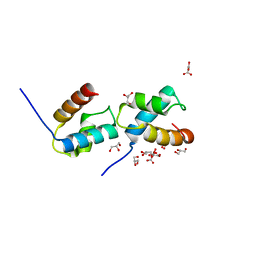 | |
6J5B
 
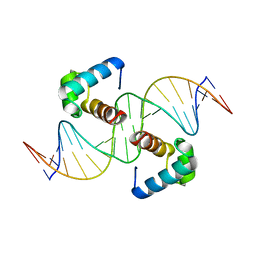 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*GP*GP*TP*AP*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*CP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*TP*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*GP*TP*AP*CP*C)-3'), Protein PHOSPHATE STARVATION RESPONSE 1 | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N, Wu, Y.K. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
