5V74
 
 | |
5ZYR
 
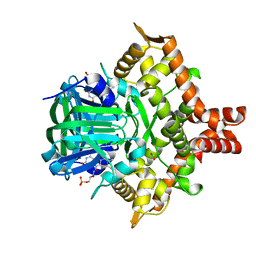 | | Crystal structure of the reductase (C1) component of p-hydroxyphenylacetate 3-hydroxylase (HPAH) from Acinetobacter baumannii | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, p-hydroxyphenylacetate 3-hydroxylase, ... | | Authors: | Oonanant, W, Phongsak, T, Sucharitakul, J, Chaiyen, P, Yuvaniyama, J. | | Deposit date: | 2018-05-28 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.20001316 Å) | | Cite: | Crystal structure of the reductase (C1) component of p-hydroxyphenylacetate 3-hydroxylase (HPAH) from Acinetobacter baumannii
To Be Published
|
|
8EUO
 
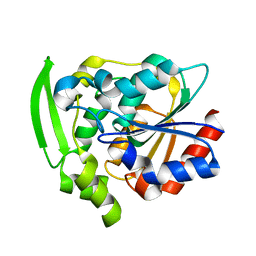 | | Hydroxynitrile Lyase from Hevea brasiliensis with Seven Mutations | | Descriptor: | (S)-hydroxynitrile lyase | | Authors: | Greenberg, L.R, Walsh, M.E, Kazlauskas, R.J, Pierce, C.T, Shi, K, Aihara, H, Evans, R.L. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | to be published
To Be Published
|
|
8ERX
 
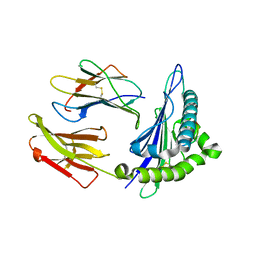 | | Structure of chimeric HLA-A*11:01-A*02:01 bound to HIV-1 RT peptide | | Descriptor: | Beta-2-microglobulin, HIV-1 RT, HLA-A*02:01 | | Authors: | Florio, T.J, Ani, O, Young, M.C, Mallik, L, Sgourakis, N.G. | | Deposit date: | 2022-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Decoupling peptide binding from T cell receptor recognition with engineered chimeric MHC-I molecules.
Front Immunol, 14, 2023
|
|
9FMX
 
 | | Aerolysin Y221G - prepore | | Descriptor: | Aerolysin | | Authors: | Iacovache, I, Zuber, B. | | Deposit date: | 2024-06-07 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Aerolysin Nanopore Structures Revealed at High Resolution in a Lipid Environment.
J.Am.Chem.Soc., 147, 2025
|
|
8EGK
 
 | | Re-refinement of Crystal Structure of NosGet3d, the All4481 protein from Nostoc sp. PCC 7120 | | Descriptor: | AZIDE ION, All4481 protein | | Authors: | Barlow, A.N, Manu, M.S, Ramasamy, S, Clemons Jr, W.M. | | Deposit date: | 2022-09-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of Get3d reveal a distinct architecture associated with the emergence of photosynthesis.
J.Biol.Chem., 299, 2023
|
|
8FSD
 
 | | P130R mutant of soybean SHMT8 in complex with PLP-glycine and formylTHF | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Beamer, L.J, Korasick, D.A. | | Deposit date: | 2023-01-09 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and functional analysis of two SHMT8 variants associated with soybean cyst nematode resistance.
Febs J., 291, 2024
|
|
9MUP
 
 | |
8V1X
 
 | | Crystal structure of polyketide synthase (PKS) thioreductase domain from Streptomyces coelicolor | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pereira, J.H, Dan, Q, Keasling, J, Adams, P.D. | | Deposit date: | 2023-11-21 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Polyketide synthase-based biosynthesis of diols and aldehyde derivatives via terminal thioreduction
To Be Published
|
|
9MUA
 
 | |
9R2O
 
 | | De novo designed N5 protein fold | | Descriptor: | N5, POTASSIUM ION, THIOCYANATE ION | | Authors: | Pacesa, M, Miao, Y, Georgeon, S, Schmidt, J, Correia, B.E. | | Deposit date: | 2025-04-30 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Reimagine accessibility of protein space with generative model
To Be Published
|
|
9R2R
 
 | |
9R2L
 
 | | De novo designed N37 protein fold | | Descriptor: | ACETATE ION, ETHANOLAMINE, N37, ... | | Authors: | Pacesa, M, Miao, Y, Georgeon, S, Schmidt, J, Correia, B.E. | | Deposit date: | 2025-04-30 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reimagine accessibility of protein space with generative model
To Be Published
|
|
9NA2
 
 | | IRAK4 in Complex with Compound 9 | | Descriptor: | (6P)-6-[(8R)-3-cyanopyrrolo[1,2-b]pyridazin-7-yl]-N-[(2R)-2-fluoro-3-hydroxy-3-methylbutyl]-4-(methylamino)pyridine-3-carboxamide, DIMETHYL SULFOXIDE, Interleukin-1 receptor-associated kinase 4 | | Authors: | Ferrao, R, Lansdon, E.B. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery of Edecesertib (GS-5718): A Potent, Selective Inhibitor of IRAK4.
J.Med.Chem., 2025
|
|
9R2V
 
 | | De novo designed M16 protein fold | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, FORMIC ACID, ... | | Authors: | Pacesa, M, Miao, Y, Georgeon, S, Schmidt, J, Correia, B.E. | | Deposit date: | 2025-05-01 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Reimagine accessibility of protein space with generative model
To Be Published
|
|
9NA3
 
 | | IRAK4 in Complex with Compound 15 | | Descriptor: | (6P)-6-[(8R)-3-cyanopyrrolo[1,2-b]pyridazin-7-yl]-N-[(2S)-2-fluoro-3-hydroxy-3-methylbutyl]-4-[(4-hydroxybicyclo[2.2.2]octan-1-yl)amino]pyridine-3-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Ferrao, R, Lansdon, E.B. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Edecesertib (GS-5718): A Potent, Selective Inhibitor of IRAK4.
J.Med.Chem., 2025
|
|
9NA4
 
 | | IRAK4 in Complex with Compound 18 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, methyl {[(1S,3s)-3-{[(2P)-2-[(8R)-3-cyanopyrrolo[1,2-b]pyridazin-7-yl]-5-{[(2S)-2-fluoro-3-hydroxy-3-methylbutyl]carbamoyl}pyridin-4-yl]amino}cyclobutyl]methyl}carbamate | | Authors: | Ferrao, R, Lansdon, E.B. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery of Edecesertib (GS-5718): A Potent, Selective Inhibitor of IRAK4.
J.Med.Chem., 2025
|
|
9NA6
 
 | | IRAK4 in Complex with Compound 34 | | Descriptor: | (6P)-4-{[(1S)-1-cyanoethyl]amino}-6-[(8S)-3-cyanopyrrolo[1,2-b]pyridazin-7-yl]-N-[(2S)-2-fluoro-3-hydroxy-3-methylbutyl]pyridine-3-carboxamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Ferrao, R, Lansdon, E.B. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of Edecesertib (GS-5718): A Potent, Selective Inhibitor of IRAK4.
J.Med.Chem., 2025
|
|
9QMP
 
 | | E.coli seryl-tRNA synthetase | | Descriptor: | Serine--tRNA ligase | | Authors: | Cusack, S. | | Deposit date: | 2025-03-24 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A.
Nature, 347, 1990
|
|
9NA5
 
 | | IRAK4 in Complex with Compound 24 | | Descriptor: | (6P)-6-[(8R)-3-cyanopyrrolo[1,2-b]pyridazin-7-yl]-4-({(1s,4S)-4-[1-(difluoromethyl)-1H-pyrazol-4-yl]cyclohexyl}amino)-N-[(2S)-2-fluoro-3-hydroxy-3-methylbutyl]pyridine-3-carboxamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Ferrao, R, Lansdon, E.B. | | Deposit date: | 2025-02-11 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of Edecesertib (GS-5718): A Potent, Selective Inhibitor of IRAK4.
J.Med.Chem., 2025
|
|
8W37
 
 | |
8W2Y
 
 | |
8W32
 
 | |
4TNJ
 
 | | RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 4.5 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4TNI
 
 | | RT XFEL structure of Photosystem II 500 ms after the third illumination at 4.6 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
