1SLT
 
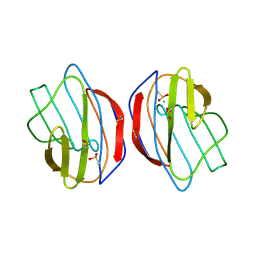 | |
1S2T
 
 | | Crystal Structure Of Apo Phosphoenolpyruvate Mutase | | Descriptor: | Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1S2U
 
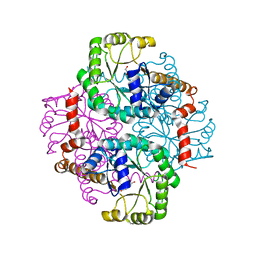 | | Crystal structure of the D58A phosphoenolpyruvate mutase mutant protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1S2W
 
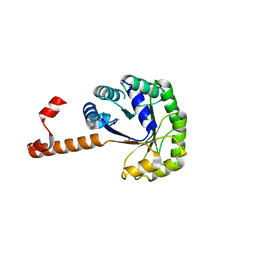 | | Crystal structure of phosphoenolpyruvate mutase in high ionic strength | | Descriptor: | Phosphoenolpyruvate phosphomutase, SULFATE ION | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1R8G
 
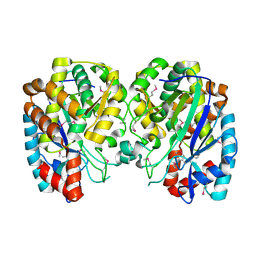 | | Structure and function of YbdK | | Descriptor: | Hypothetical protein ybdK | | Authors: | Lehmann, C, Doseeva, V, Pullalarevu, S, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-10-24 | | Release date: | 2004-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
1S2V
 
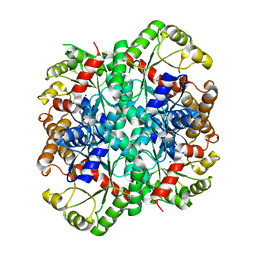 | | Crystal structure of phosphoenolpyruvate mutase complexed with Mg(II) | | Descriptor: | MAGNESIUM ION, Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1RXX
 
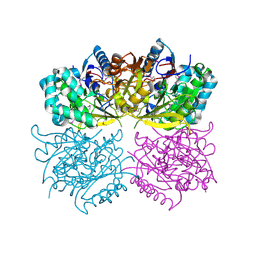 | | Structure of arginine deiminase | | Descriptor: | Arginine deiminase | | Authors: | Galkin, A, Kulakova, L, Sarikaya, E, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insight into arginine degradation by arginine deiminase, an antibacterial and parasite drug target.
J.Biol.Chem., 279, 2004
|
|
1RW1
 
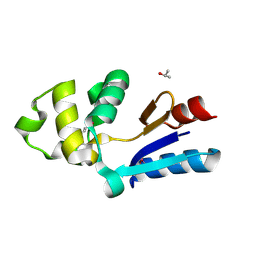 | | YFFB (PA3664) PROTEIN | | Descriptor: | ISOPROPYL ALCOHOL, conserved hypothetical protein yffB | | Authors: | Teplyakov, A, Pullalarevu, S, Obmolova, G, Doseeva, V, Galkin, A, Herzberg, O, Dauter, M, Dauter, Z, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-12-15 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the YffB protein from Pseudomonas aeruginosa suggests a glutathione-dependent thiol reductase function.
Bmc Struct.Biol., 4, 2004
|
|
1F3G
 
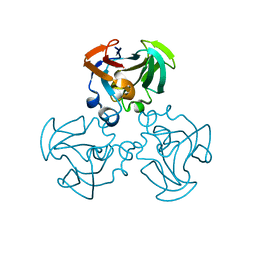 | | THREE-DIMENSIONAL STRUCTURE OF THE ESCHERICHIA COLI PHOSPHOCARRIER PROTEIN III GLC | | Descriptor: | GLUCOSE-SPECIFIC PHOSPHOCARRIER PROTEIN IIAGLC | | Authors: | Worthylake, D, Meadow, N, Roseman, S, Liao, D.-I, Herzberg, O, Remington, S.J. | | Deposit date: | 1991-08-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of the Escherichia coli phosphocarrier protein IIIglc.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1DBU
 
 | | Crystal structure of cysteinyl-tRNA(Pro) deacylase protein from H. influenzae (HI1434) | | Descriptor: | MERCURY (II) ION, cysteinyl-tRNA(Pro) deacylase | | Authors: | Zhang, H, Huang, K, Li, Z, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 1999-11-03 | | Release date: | 2000-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.
Proteins, 40, 2000
|
|
1DBX
 
 | | Crystal structure of cysteinyl-tRNA(Pro) deacylase from H. influenzae (HI1434) | | Descriptor: | cysteinyl-tRNA(Pro) deacylase | | Authors: | Zhang, H, Huang, K, Li, Z, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 1999-11-03 | | Release date: | 2000-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.
Proteins, 40, 2000
|
|
1M1B
 
 | | Crystal Structure of Phosphoenolpyruvate Mutase Complexed with Sulfopyruvate | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE PHOSPHOMUTASE, SULFOPYRUVATE | | Authors: | Liu, S, Lu, Z, Jia, Y, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2002-06-18 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dissociative phosphoryl transfer in PEP mutase catalysis: structure of the enzyme/sulfopyruvate complex and kinetic properties of mutants.
Biochemistry, 41, 2002
|
|
1IZM
 
 | | Structure of ygfB from Haemophilus influenzae (HI0817), a Conserved Hypothetical Protein | | Descriptor: | HYPOTHETICAL PROTEIN HI0817 | | Authors: | Galkin, A, Sarikaya, E, Lehmann, C, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-10-09 | | Release date: | 2003-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray structure of HI0817 from Haemophilus influenzae: protein of unknown function with a novel fold.
Proteins, 57, 2004
|
|
1J7G
 
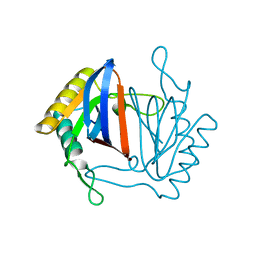 | |
1J8B
 
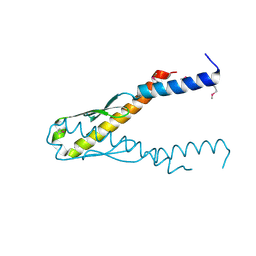 | | Structure of YbaB from Haemophilus influenzae (HI0442), a protein of unknown function | | Descriptor: | YbaB | | Authors: | Lim, K, Tempcyzk, A, Toedt, J, Parsons, J.F, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-21 | | Release date: | 2003-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of YbaB from Haemophilus influenzae (HI0442), a
protein of unknown function coexpressed with the recombinational
DNA repair protein RecR
Proteins, 50, 2003
|
|
1IM8
 
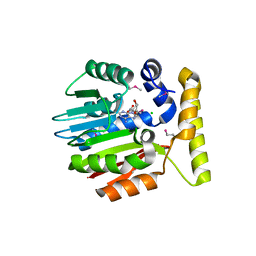 | | Crystal structure of YecO from Haemophilus influenzae (HI0319), a methyltransferase with a bound S-adenosylhomocysteine | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOSELENOCYSTEINE, YecO | | Authors: | Lim, K, Zhang, H, Tempczyk, A, Bonander, N, Toedt, J, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-10 | | Release date: | 2001-11-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of YecO from Haemophilus influenzae (HI0319) reveals a methyltransferase fold and a bound S-adenosylhomocysteine.
Proteins, 45, 2001
|
|
1MWQ
 
 | | Structure of HI0828, a Hypothetical Protein from Haemophilus influenzae with a Putative Active-Site Phosphohistidine | | Descriptor: | CACODYLATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Willis, M.A, Krajewski, W, Chalamasetty, V.R, Reddy, P, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-09-30 | | Release date: | 2003-11-25 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structure of YciI from Haemophilus influenzae (HI0828) reveals a ferredoxin-like alpha/beta-fold with a histidine/aspartate centered catalytic site
Proteins, 59, 2005
|
|
1J85
 
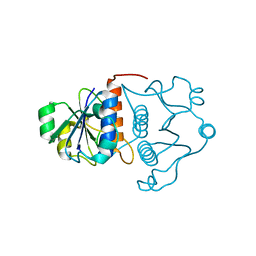 | | Structure of YibK from Haemophilus influenzae (HI0766), a truncated sequence homolog of tRNA (guanosine-2'-O-) methyltransferase (SpoU) | | Descriptor: | YibK | | Authors: | Lim, K, Zhang, H, Toedt, J, Tempcyzk, A, Krajewski, W, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-20 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the YibK methyltransferase from Haemophilus influenzae
(HI0766): A cofactor bound at a site formed by a knot
Proteins, 51, 2003
|
|
1J8D
 
 | |
1NNX
 
 | | Structure of the hypothetical protein ygiW from E. coli. | | Descriptor: | Protein ygiW, SULFATE ION | | Authors: | Lehmann, C, Galkin, A, Pullalarevu, S, Sarikaya, E, Krajewski, W, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-14 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the hypothetical protein ygiW from E. coli.
To be Published
|
|
1MW5
 
 | | Structure of HI1480 from Haemophilus influenzae | | Descriptor: | HYPOTHETICAL PROTEIN HI1480 | | Authors: | Lim, K, Sarikaya, E, Howard, A, Galkin, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-09-27 | | Release date: | 2003-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel structure and nucleotide binding properties of HI1480 from Haemophilus influenzae: a protein with no known sequence homologues
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
1JN1
 
 | | Structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Haemophilus influenzae (HI0671) | | Descriptor: | 2C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE SYNTHASE, COBALT (II) ION, SULFATE ION | | Authors: | Lehmann, C, Lim, K, Toedt, J, Krajewski, W, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-21 | | Release date: | 2002-08-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of 2C-methyl-D-erythrol-2,4-cyclodiphosphate synthase from Haemophilus influenzae: activation by conformational transition.
Proteins, 49, 2002
|
|
1JO0
 
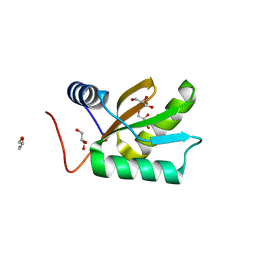 | | Structure of HI1333, a Hypothetical Protein from Haemophilus influenzae with Structural Similarity to RNA-binding Proteins | | Descriptor: | GLYCEROL, HYPOTHETICAL PROTEIN HI1333 | | Authors: | Willis, M.A, Krajewski, W, Chalamasetty, V.R, Reddy, P, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-26 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of HI1333 (YhbY), a putative RNA-binding protein from Haemophilus influenzae
Proteins, 49, 2002
|
|
1OQF
 
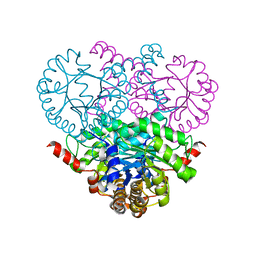 | | Crystal structure of the 2-methylisocitrate lyase | | Descriptor: | 2-methylisocitrate lyase | | Authors: | Liu, S, Lu, Z, Dunaway-Mariano, D, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-03-08 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 2-methylisocitrate lyase in complex with product and with isocitrate inhibitor provide insight into lyase substrate specificity, catalysis and evolution.
Biochemistry, 44, 2005
|
|
1K1E
 
 | |
