5WI8
 
 | |
5WIW
 
 | |
7U4Z
 
 | |
5USH
 
 | | Structure of vaccinia virus D8 protein bound to human Fab vv66 | | Descriptor: | Fab vv66 heavy chain, Fab vv66 light chain, IMV membrane protein | | Authors: | Zajonc, D.M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function characterization of three human antibodies targeting the vaccinia virus adhesion molecule D8.
J. Biol. Chem., 293, 2018
|
|
5USL
 
 | | Structure of vaccinia virus D8 protein bound to human Fab vv304 | | Descriptor: | Fab vv304 Heavy chain, Fab vv304 Light chain, IMV membrane protein | | Authors: | Zajonc, D.M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function characterization of three human antibodies targeting the vaccinia virus adhesion molecule D8.
J. Biol. Chem., 293, 2018
|
|
9DP8
 
 | | A DARPin fused to the 1TEL crystallization chaperone via a helical Lys-Gln-Arg linker | | Descriptor: | Transcription Factor ETV6, DARPin fusion | | Authors: | Pedroza Romo, M.J, Keliiliki, A.C, Noakes, E, Averett, B, Moody, J.D. | | Deposit date: | 2024-09-20 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Optimal TELSAM-Target Protein Linker Character is Target Protein Dependent.
To Be Published
|
|
6B9J
 
 | | Structure of vaccinia virus D8 protein bound to human Fab vv138 | | Descriptor: | Fab vv138 Heavy chain, Fab vv138 Light chain, GLYCEROL, ... | | Authors: | Zajonc, D.M. | | Deposit date: | 2017-10-10 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function characterization of three human antibodies targeting the vaccinia virus adhesion molecule D8.
J. Biol. Chem., 293, 2018
|
|
7TDY
 
 | | The ubiquitin-associated domain of human thirty-eight negative kinase 1, flexibly fused to the 1TEL crystallization chaperone via a 2-glycine linker and crystallized at low protein concentration | | Descriptor: | FORMIC ACID, Transcription factor ETV6,Non-receptor tyrosine-protein kinase TNK1 | | Authors: | Nawarathnage, S, Bunn, D.R, Stewart, C, Doukev, T, Moody, J.D. | | Deposit date: | 2022-01-03 | | Release date: | 2023-01-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Fusion crystallization reveals the behavior of both the 1TEL crystallization chaperone and the TNK1 UBA domain.
Structure, 31, 2023
|
|
6UCW
 
 | | Multi-conformer model of Apo Ketosteroid Isomerase from Pseudomonas Putida (pKSI) at 250 K | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Steroid Delta-isomerase | | Authors: | Yabukarski, F, Herschlag, D, Biel, J.T, Fraser, J.S. | | Deposit date: | 2019-09-17 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UBQ
 
 | |
6TZD
 
 | |
6U1Z
 
 | |
6U4I
 
 | |
6UCN
 
 | | Multi-conformer model of Ketosteroid Isomerase from Pseudomonas Putida (pKSI) bound to Equilenin at 250 K | | Descriptor: | CHLORIDE ION, EQUILENIN, MAGNESIUM ION, ... | | Authors: | Yabukarski, F, Herschlag, D, Biel, J.T, Fraser, J.S. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5TOO
 
 | | Crystal structure of alkaline phosphatase PafA T79S, N100A, K162A, R164A mutant | | Descriptor: | Alkaline phosphatase PafA, CHLORIDE ION, ZINC ION | | Authors: | Lyubimov, A.Y, Sunden, F, AlSadhan, I, Herschlag, D. | | Deposit date: | 2016-10-18 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Differential catalytic promiscuity of the alkaline phosphatase superfamily bimetallo core reveals mechanistic features underlying enzyme evolution.
J. Biol. Chem., 292, 2017
|
|
6CPR
 
 | | Crystal structure of 4-1BBL/4-1BB complex in C2 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Aruna, B, Zajonc, D.M. | | Deposit date: | 2018-03-14 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the human 4-1BB receptor bound to its ligand 4-1BBL reveal covalent receptor dimerization as a potential signaling amplifier.
J. Biol. Chem., 293, 2018
|
|
6C1X
 
 | | Crystal Structure of Ketosteroid Isomerase D40N/D103N mutant from Pseudomonas Putida (pKSI) bound to 3,4-dinitrophenol | | Descriptor: | 3,4-dinitrophenol, MAGNESIUM ION, Steroid Delta-isomerase | | Authors: | Yabukarski, F, Pinney, M.M, Herschlag, D. | | Deposit date: | 2018-01-05 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Coupling Throughout the Active Site Hydrogen Bond Networks of Ketosteroid Isomerase and Photoactive Yellow Protein.
J. Am. Chem. Soc., 140, 2018
|
|
6C17
 
 | | Crystal Structure of Ketosteroid Isomerase D40N mutant from Pseudomonas Putida (pKSI) bound to 3,4-dinitrophenol | | Descriptor: | 3,4-dinitrophenol, MAGNESIUM ION, Steroid Delta-isomerase | | Authors: | Yabukarski, F, Pinney, M, Herschlag, D. | | Deposit date: | 2018-01-04 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Coupling Throughout the Active Site Hydrogen Bond Networks of Ketosteroid Isomerase and Photoactive Yellow Protein.
J. Am. Chem. Soc., 140, 2018
|
|
6C1J
 
 | | Crystal Structure of Ketosteroid Isomerase Y32F/Y57F/D40N mutant from Pseudomonas Putida (pKSI) bound to 3,4-dinitrophenol | | Descriptor: | 3,4-dinitrophenol, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Yabukarski, F, Pinney, M.M, Herschlag, D. | | Deposit date: | 2018-01-04 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.063 Å) | | Cite: | Structural Coupling Throughout the Active Site Hydrogen Bond Networks of Ketosteroid Isomerase and Photoactive Yellow Protein.
J. Am. Chem. Soc., 140, 2018
|
|
3JT8
 
 | | Structure of neuronal nitric oxide synthase heme domain complexed with N~5~-{3-[(1-methylethyl)sulfanyl]propanimidoyl}-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Heme-coordinating inhibitors of neuronal nitric oxide synthase. Iron-thioether coordination is stabilized by hydrophobic contacts without increased inhibitor potency.
J.Am.Chem.Soc., 132, 2010
|
|
3JTA
 
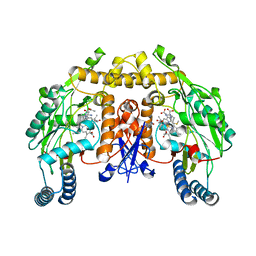 | | Structure of neuronal nitric oxide synthase heme domain in the ferrous state complexed with N~5~-[4-(methylsulfanyl)butanimidoyl]-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Heme-coordinating inhibitors of neuronal nitric oxide synthase. Iron-thioether coordination is stabilized by hydrophobic contacts without increased inhibitor potency.
J.Am.Chem.Soc., 132, 2010
|
|
3JT4
 
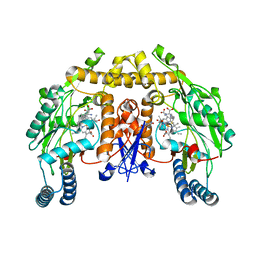 | | Structure of neuronal nitric oxide synthase heme domain complexed with N~5~-[(3-(ethylsulfanyl)propanimidoyl]-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Heme-coordinating inhibitors of neuronal nitric oxide synthase. Iron-thioether coordination is stabilized by hydrophobic contacts without increased inhibitor potency.
J.Am.Chem.Soc., 132, 2010
|
|
3JT7
 
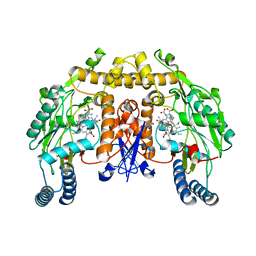 | | Structure of neuronal nitric oxide synthase heme domain complexed with N~5~-[2-(propylsulfanyl)ethanimidoyl]-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Heme-coordinating inhibitors of neuronal nitric oxide synthase. Iron-thioether coordination is stabilized by hydrophobic contacts without increased inhibitor potency.
J.Am.Chem.Soc., 132, 2010
|
|
3JT9
 
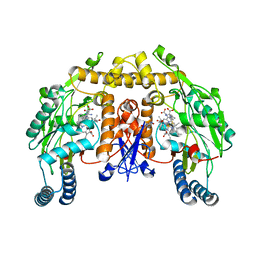 | | Structure of neuronal nitric oxide synthase heme domain in the ferrous state complexed with N~5~-[2-(ethylsulfanyl)ethanimidoyl]-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Heme-coordinating inhibitors of neuronal nitric oxide synthase. Iron-thioether coordination is stabilized by hydrophobic contacts without increased inhibitor potency.
J.Am.Chem.Soc., 132, 2010
|
|
3JT5
 
 | | Structure of neuronal nitric oxide synthase heme domain complexed with N~5~-[2-(ethylsulfanyl)ethanimidoyl]-L-ornithine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Heme-coordinating inhibitors of neuronal nitric oxide synthase. Iron-thioether coordination is stabilized by hydrophobic contacts without increased inhibitor potency.
J.Am.Chem.Soc., 132, 2010
|
|
