6VCX
 
 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 1 (AtMAT1) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
6VD1
 
 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 2 (AtMAT2) in complex with S-adenosylmethionine and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
6VD0
 
 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 2 (AtMAT2) in complex with free Methionine and AMPCPP | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
6BST
 
 | |
6VCY
 
 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 1 (AtMAT1) in complex with 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
6AQV
 
 | | Crystal Structure of Z-DNA with 6-fold Twinning_Z3B | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Luo, Z, Dauter, Z, Gilski, M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Four highly pseudosymmetric and/or twinned structures of d(CGCGCG)2 extend the repertoire of crystal structures of Z-DNA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6VCZ
 
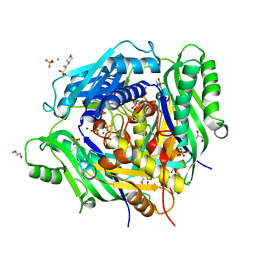 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 2 (AtMAT2) | | Descriptor: | 2-METHOXYETHANOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
6AQT
 
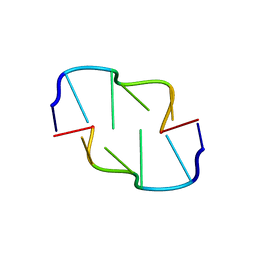 | | Crystal Structure of Z-DNA with 6-fold Twinning_Z3A | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Luo, Z, Dauter, Z, Gilski, M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Four highly pseudosymmetric and/or twinned structures of d(CGCGCG)2 extend the repertoire of crystal structures of Z-DNA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6AQW
 
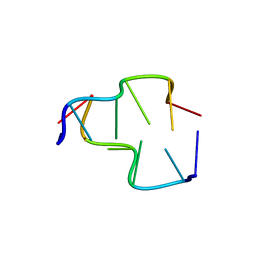 | | Crystal Structure of Z-DNA with 6-fold Twinning_Z4A | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Luo, Z, Dauter, Z, Gilski, M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Four highly pseudosymmetric and/or twinned structures of d(CGCGCG)2 extend the repertoire of crystal structures of Z-DNA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6AQX
 
 | | Crystal Structure of Z-DNA with 6-fold Twinning_Z4B | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Luo, Z, Dauter, Z, Gilski, M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Four highly pseudosymmetric and/or twinned structures of d(CGCGCG)2 extend the repertoire of crystal structures of Z-DNA.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1RSN
 
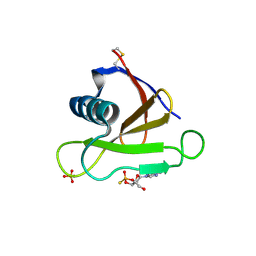 | | RIBONUCLEASE (RNASE SA) (E.C.3.1.4.8) COMPLEXED WITH EXO-2',3'-CYCLOPHOSPHOROTHIOATE | | Descriptor: | GUANOSINE-2',3'-CYCLOPHOSPHOROTHIOATE, RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Dauter, Z, Lamzin, V.S, Wilson, K.S. | | Deposit date: | 1995-09-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex of ribonuclease Sa with a cyclic nucleotide and a proposed model for the reaction intermediate.
Eur.J.Biochem., 216, 1993
|
|
3HQX
 
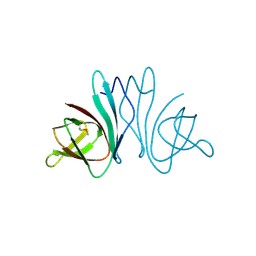 | | Crystal structure of protein of unknown function (DUF1255,PF06865) from Acinetobacter sp. ADP1 | | Descriptor: | UPF0345 protein ACIAD0356 | | Authors: | Nocek, B, Hatzos, C, Freeman, L, Dauter, Z, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-08 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of protein of unknown function (DUF1255,PF06865) from Acinetobacter sp. ADP1
To be Published
|
|
3IO2
 
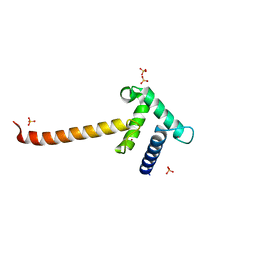 | | Crystal structure of the Taz2 domain of p300 | | Descriptor: | Histone acetyltransferase p300, SULFATE ION, ZINC ION | | Authors: | Miller, M, Dauter, Z, Wlodawer, A. | | Deposit date: | 2009-08-13 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Taz2 domain of p300: insights into ligand binding.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3H1R
 
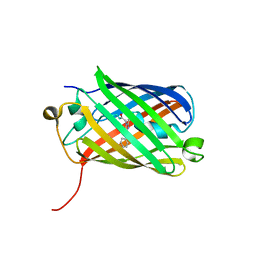 | | Order-disorder structure of fluorescent protein FP480 | | Descriptor: | Fluorescent protein FP480 | | Authors: | Pletnev, S, Morozova, K.S, Verkhusha, V.V, Dauter, Z. | | Deposit date: | 2009-04-13 | | Release date: | 2009-09-08 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Rotational order-disorder structure of fluorescent protein FP480
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3H1O
 
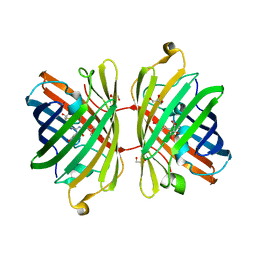 | | The Structure of Fluorescent Protein FP480 | | Descriptor: | Fluorescent protein FP480, GLYCEROL | | Authors: | Pletnev, S, Morozova, K.S, Verkhusha, V.V, Dauter, Z. | | Deposit date: | 2009-04-13 | | Release date: | 2009-09-08 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rotational order-disorder structure of fluorescent protein FP480
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4Q7R
 
 | |
5BSG
 
 | | Crystal structure of Medicago truncatula (delta)1-Pyrroline-5-Carboxylate Reductase (MtP5CR) in complex with NADP+ | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Ruszkowski, M, Nocek, B, Forlani, G, Dauter, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of Medicago truncatula delta (1)-pyrroline-5-carboxylate reductase provides new insights into regulation of proline biosynthesis in plants.
Front Plant Sci, 6, 2015
|
|
5BSE
 
 | | Crystal structure of Medicago truncatula (delta)1-Pyrroline-5-Carboxylate Reductase (MtP5CR) | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, Pyrroline-5-carboxylate reductase | | Authors: | Ruszkowski, M, Nocek, B, Forlani, G, Dauter, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of Medicago truncatula delta (1)-pyrroline-5-carboxylate reductase provides new insights into regulation of proline biosynthesis in plants.
Front Plant Sci, 6, 2015
|
|
5BSH
 
 | | Crystal structure of Medicago truncatula (delta)1-Pyrroline-5-Carboxylate Reductase (MtP5CR) in complex with L-Proline | | Descriptor: | PROLINE, Pyrroline-5-carboxylate reductase | | Authors: | Ruszkowski, M, Nocek, B, Forlani, G, Dauter, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of Medicago truncatula delta (1)-pyrroline-5-carboxylate reductase provides new insights into regulation of proline biosynthesis in plants.
Front Plant Sci, 6, 2015
|
|
5BSF
 
 | | Crystal structure of Medicago truncatula (delta)1-Pyrroline-5-Carboxylate Reductase (MtP5CR) in complex with NAD+ | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Ruszkowski, M, Nocek, B, Forlani, G, Dauter, Z. | | Deposit date: | 2015-06-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of Medicago truncatula delta (1)-pyrroline-5-carboxylate reductase provides new insights into regulation of proline biosynthesis in plants.
Front Plant Sci, 6, 2015
|
|
352D
 
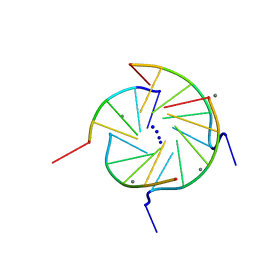 | | THE CRYSTAL STRUCTURE OF A PARALLEL-STRANDED PARALLEL-STRANDED GUANINE TETRAPLEX AT 0.95 ANGSTROM RESOLUTION | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3'), SODIUM ION | | Authors: | Phillips, K, Dauter, Z, Murchie, A.I.H, Lilley, D.M.J, Luisi, B. | | Deposit date: | 1997-09-04 | | Release date: | 1997-11-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The crystal structure of a parallel-stranded guanine tetraplex at 0.95 A resolution.
J.Mol.Biol., 273, 1997
|
|
310D
 
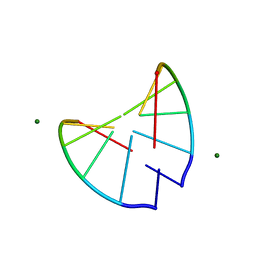 | | Crystal structure of 2'-O-Me(CGCGCG)2: an RNA duplex at 1.3 A resolution. Hydration pattern of 2'-O-methylated RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(OMC)P*(OMG)P*(OMC)P*(OMG)P*(OMC)P*(OMG))-3') | | Authors: | Adamiak, D.A, Milecki, J, Adamiak, R.W, Dauter, Z, Rypniewski, W.R. | | Deposit date: | 1996-05-25 | | Release date: | 1997-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of 2'-O-Me(CGCGCG)2, an RNA duplex at 1.30 A resolution. Hydration pattern of 2'-O-methylated RNA.
Nucleic Acids Res., 25, 1997
|
|
5EMG
 
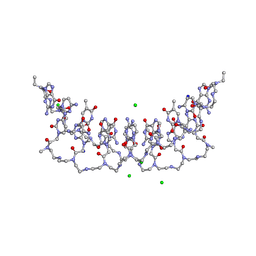 | | Crystal structures of PNA p(GCTGCTGC)2 duplex containing T-T mismatches | | Descriptor: | CHLORIDE ION, GPN-CPN-TPN-GPN-CPN-TPN-GPN-CPN, SODIUM ION | | Authors: | Kiliszek, A, Banaszak, K, Dauter, Z, Rypniewski, W. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | The first crystal structures of RNA-PNA duplexes and a PNA-PNA duplex containing mismatches-toward anti-sense therapy against TREDs.
Nucleic Acids Res., 44, 2016
|
|
5EQ8
 
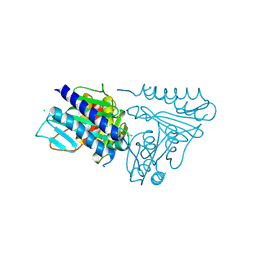 | |
5EMF
 
 | | Crystal structure of RNA r(GCUGCUGC) with antisense PNA p(GCAGCAGC) | | Descriptor: | CHLORIDE ION, RNA (5'-R(*GP*CP*UP*GP*CP*UP*GP*C)-3'), antisense PNA p(GCAGCAGC) | | Authors: | Kiliszek, A, Banaszak, K, Dauter, Z, Rypniewski, W. | | Deposit date: | 2015-11-06 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | The first crystal structures of RNA-PNA duplexes and a PNA-PNA duplex containing mismatches-toward anti-sense therapy against TREDs.
Nucleic Acids Res., 44, 2016
|
|
