3ROZ
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein in Complex with Nicotinamide | | Descriptor: | Apolipoprotein A-I-binding protein, NICOTINAMIDE, SULFATE ION | | Authors: | Shumilin, I.A, Jha, K.N, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2011-04-26 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RQ8
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis soaked with P1,P5-Di(adenosine-5') pentaphosphate | | Descriptor: | ADP/ATP-dependent NAD(P)H-hydrate dehydratase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RU3
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADPH and ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BETA-6-HYDROXY-1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-04 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RU2
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADPH. | | Descriptor: | BETA-6-HYDROXY-1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-04 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
4TNN
 
 | | Crystal structure of Escherichia coli protein YodA in complex with Ni - artifact of purification. | | Descriptor: | Metal-binding lipocalin, NICKEL (II) ION, SULFATE ION | | Authors: | Gasiorowska, O.A, Cymborowski, M.T, Handing, K.B, Shabalin, I.G, Zasadzinska, E, Niedzialkowska, E, Porebski, P.J, Minor, W. | | Deposit date: | 2014-06-04 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
4YYC
 
 | | Crystal structure of trimethylamine methyltransferase from Sinorhizobium meliloti in complex with unknown ligand | | Descriptor: | CHLORIDE ION, Putative trimethylamine methyltransferase, UNKNOWN LIGAND | | Authors: | Shabalin, I.G, Porebski, P.J, Gasiorowska, O.A, Handing, K.B, Niedzialkowska, E, Cymborowski, M.T, Cooper, D.R, Stead, M, Hammonds, J, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
4ZNZ
 
 | | Crystal structure of Escherichia coli carbonic anhydrase (YadF) in complex with Zn - artifact of purification | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Gasiorowska, O.A, Niedzialkowska, E, Porebski, P.J, Handing, K.B, Shabalin, I.G, Cymborowski, M.T, Minor, W. | | Deposit date: | 2015-05-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
6CXD
 
 | | Crystal structure of peptidase B from Yersinia pestis CO92 at 2.75 A resolution | | Descriptor: | Peptidase B, SULFATE ION | | Authors: | Woinska, M, Lipowska, J, Shabalin, I.G, Cymborowski, M, Grimshaw, S, Winsor, J, Shuvalova, L, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-02 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
6HCD
 
 | | Structure of universal stress protein from Archaeoglobus fulgidus | | Descriptor: | ACETATE ION, CHLORIDE ION, UNIVERSAL STRESS PROTEIN, ... | | Authors: | Shumilin, I.A, Loch, J.I, Cymborowski, M, Xu, X, Edwards, A, Di Leo, R, Shabalin, I.G, Joachimiak, A, Savchenko, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-08-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
3CNI
 
 | | Crystal structure of a domain of a putative ABC type-2 transporter from Thermotoga maritima MSB8 | | Descriptor: | CALCIUM ION, Putative ABC type-2 transporter | | Authors: | Filippova, E.V, Shumilin, I, Tkaczuk, K.L, Cymborowski, M, Chruszcz, M, Xu, X, Que, Q, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of the putative ABC-type 2 transporter from Thermotoga maritima MSB8.
J.Struct.Funct.Genom., 15, 2014
|
|
7SBG
 
 | |
7SD2
 
 | |
3MLE
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori cocrystallized with ATP | | Descriptor: | 8-aminooctanoic acid, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Nicholls, R, Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-16 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
2IS3
 
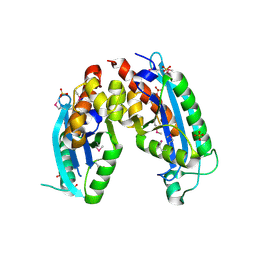 | |
3FTT
 
 | | Crystal Structure of the galactoside O-acetyltransferase from Staphylococcus aureus | | Descriptor: | Putative acetyltransferase SACOL2570 | | Authors: | Knapik, A.A, Shumilin, I.A, Cui, H, Xu, X, Chruszcz, M, Zimmerman, M.D, Cymborowski, M, Anderson, W.F, Savchenko, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-13 | | Release date: | 2009-03-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biophysical analysis of the putative acetyltransferase SACOL2570 from methicillin-resistant Staphylococcus aureus.
J.Struct.Funct.Genom., 14, 2013
|
|
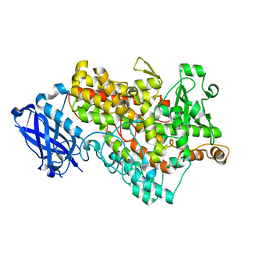
1FGT
 
 | |
1F8N
 
 | |
1FGM
 
 | |
1FGQ
 
 | |
1FGR
 
 | |
1FGO
 
 | |
2O5U
 
 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- orthorhombic form (C222). | | Descriptor: | Thioesterase | | Authors: | Chruszcz, M, Wang, S, Evdokimova, E, Koclega, K.D, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
3V4E
 
 | | Crystal Structure of the galactoside O-acetyltransferase in complex with CoA | | Descriptor: | COENZYME A, DI(HYDROXYETHYL)ETHER, Galactoside O-acetyltransferase, ... | | Authors: | Knapik, A.A, Shumilin, I.A, Luo, H.-B, Chruszcz, M, Zimmerman, M.D, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biophysical analysis of the putative acetyltransferase SACOL2570 from methicillin-resistant Staphylococcus aureus.
J.Struct.Funct.Genom., 14, 2013
|
|
5C5I
 
 | | Crystal structure of NADP-dependent dehydrogenase from Rhodobacter sphaeroides | | Descriptor: | NADP-dependent dehydrogenase | | Authors: | Kowiel, M, Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Porebski, P.J, Cymborowski, M, Al Obaidi, N.F, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-19 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of NADP-dependent dehydrogenase from Rhodobacter sphaeroides
to be published
|
|
4M3S
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Xu, X, Cymborowski, M, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-08-06 | | Release date: | 2013-08-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Double trouble-Buffer selection and His-tag presence may be responsible for nonreproducibility of biomedical experiments.
Protein Sci., 23, 2014
|
|
