2QHN
 
 | | Crystal Structure of Chek1 in Complex with Inhibitor 1a | | Descriptor: | 5-ETHYL-3-METHYL-1,5-DIHYDRO-4H-PYRAZOLO[4,3-C]QUINOLIN-4-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Munshi, S. | | Deposit date: | 2007-07-02 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of a pyrazoloquinolinone class of Chk1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7DWC
 
 | | Bacteroides thetaiotaomicron VPI5482 BTAxe1 | | Descriptor: | Xylanase | | Authors: | Wang, L.Y, Wang, Y.L, Xin, F.J, Sun, L.C. | | Deposit date: | 2021-01-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Rational Design for Broadened Substrate Specificity and Enhanced Activity of a Novel Acetyl Xylan Esterase from Bacteroides thetaiotaomicron.
J.Agric.Food Chem., 69, 2021
|
|
2Q2Y
 
 | | Crystal Structure of KSP in complex with Inhibitor 1 | | Descriptor: | 1-{(3R,3AR)-3-[3-(4-ACETYLPIPERAZIN-1-YL)PROPYL]-8-FLUORO-3-PHENYL-3A,4-DIHYDRO-3H-PYRAZOLO[5,1-C][1,4]BENZOXAZIN-2-YL}ETHANONE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2007-05-29 | | Release date: | 2007-09-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 8: Design and synthesis of 1,4-diaryl-4,5-dihydropyrazoles as potent inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2SAS
 
 | |
1WWA
 
 | | NGF BINDING DOMAIN OF HUMAN TRKA RECEPTOR | | Descriptor: | PROTEIN (NERVE GROWTH FACTOR RECEPTOR TRKA) | | Authors: | Wiesmann, C, Ultsch, M.H, Bass, S.H, De Vos, A.M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the neurotrophin-binding domain of TrkA, TrkB and TrkC.
J.Mol.Biol., 290, 1999
|
|
5ZO4
 
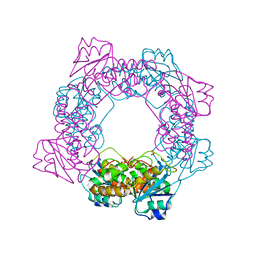 | | inactive state of the nuclease | | Descriptor: | MANGANESE (II) ION, Putative 3'-5' exonuclease family protein, SULFATE ION | | Authors: | Yuan, Z.L, Gu, L.C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
5ZO3
 
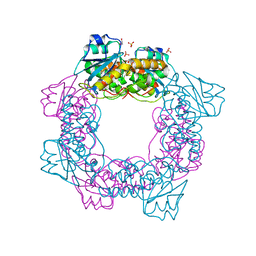 | | apo form of the nuclease | | Descriptor: | 1,2-ETHANEDIOL, Putative 3'-5' exonuclease family protein, SULFATE ION | | Authors: | Yuan, Z.L, Gu, L.C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.493 Å) | | Cite: | NrnC, an RNase D-Like Protein FromAgrobacterium, Is a Novel Octameric Nuclease That Specifically Degrades dsDNA but Leaves dsRNA Intact.
Front Microbiol, 9, 2018
|
|
1UIP
 
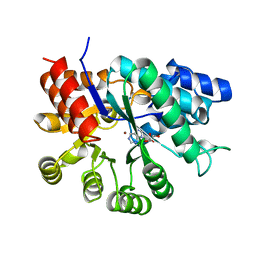 | | ADENOSINE DEAMINASE (HIS 238 GLU MUTANT) | | Descriptor: | ADENOSINE DEAMINASE, PURINE RIBOSIDE, ZINC ION | | Authors: | Wilson, D.K, Quiocho, F.A. | | Deposit date: | 1996-08-30 | | Release date: | 1997-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Site-directed mutagenesis of histidine 238 in mouse adenosine deaminase: substitution of histidine 238 does not impede hydroxylate formation.
Biochemistry, 35, 1996
|
|
2RHK
 
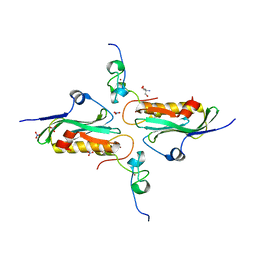 | | Crystal structure of influenza A NS1A protein in complex with F2F3 fragment of human cellular factor CPSF30, Northeast Structural Genomics Targets OR8C and HR6309A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cleavage and polyadenylation specificity factor subunit 4, NITRATE ION, ... | | Authors: | Das, K, Ma, L.-C, Xiao, R, Radvansky, B, Aramini, J, Zhao, L, Arnold, E, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-09 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for suppression of a host antiviral response by influenza A virus.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2TSA
 
 | | AZURIN MUTANT M121A | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
3CJO
 
 | | Crystal structure of KSP in complex with inhibitor 30 | | Descriptor: | (2S)-4-(2,5-difluorophenyl)-N-[(3R,4S)-3-fluoro-1-methylpiperidin-4-yl]-2-(hydroxymethyl)-N-methyl-2-phenyl-2,5-dihydro-1H-pyrrole-1-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2008-03-13 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. 9. Discovery of (2S)-4-(2,5-difluorophenyl)-n-[(3R,4S)-3-fluoro-1-methylpiperidin-4-yl]-2-(hydroxymethyl)-N-methyl-2-phenyl-2,5-dihydro-1H-pyrrole-1-carboxamide (MK-0731) for the treatment of taxane-refractory cancer.
J.Med.Chem., 51, 2008
|
|
3C1K
 
 | | Crystal structure of thrombin in complex with inhibitor 15 | | Descriptor: | 2-{3-[(benzylsulfonyl)amino]-6-methyl-2-oxopyridin-1(2H)-yl}-N-({1-[2-(tert-butylamino)-2-oxoethyl]-4-methyl-1H-imidazol-5-yl}methyl)acetamide, Hirugen, Prothrombin | | Authors: | Yan, Y. | | Deposit date: | 2008-01-23 | | Release date: | 2008-03-18 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-based design of novel groups for use in the P1 position of thrombin inhibitor scaffolds. Part 2: N-acetamidoimidazoles.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CSJ
 
 | | Human glutathione s-transferase p1-1 in complex with chlorambucil | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORAMBUCIL, CHLORIDE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The anti-cancer drug chlorambucil as a substrate for the human polymorphic enzyme glutathione transferase P1-1: kinetic properties and crystallographic characterisation of allelic variants.
J.Mol.Biol., 380, 2008
|
|
3BJT
 
 | | Pyruvate kinase M2 is a phosphotyrosine binding protein | | Descriptor: | MAGNESIUM ION, OXALATE ION, Pyruvate kinase isozymes M1/M2 | | Authors: | Wu, N. | | Deposit date: | 2007-12-04 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyruvate kinase M2 is a phosphotyrosine-binding protein.
Nature, 452, 2008
|
|
3BJF
 
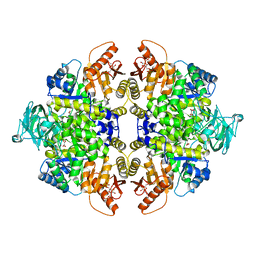 | | Pyruvate kinase M2 is a phosphotyrosine binding protein | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Wu, N. | | Deposit date: | 2007-12-03 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Pyruvate kinase M2 is a phosphotyrosine-binding protein.
Nature, 452, 2008
|
|
3CSI
 
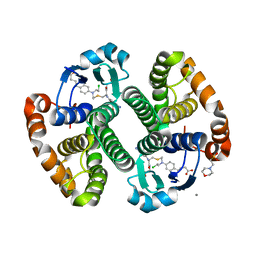 | |
3CSH
 
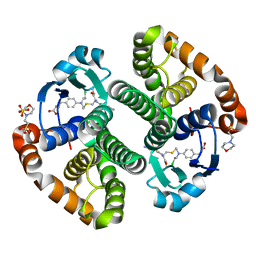 | |
3ARG
 
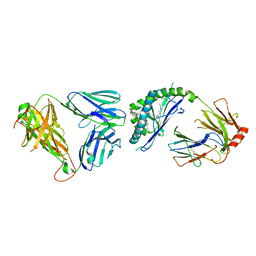 | | Ternary crystal structure of the mouse NKT TCR-CD1d-alpha-glucosylceramide(C20:2) | | Descriptor: | (11E,14E)-N-[(2S,3S,4R)-1-(alpha-D-glucopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]icosa-11,14-dienamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wun, K.S, Rossjohn, J. | | Deposit date: | 2010-11-27 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells
Immunity, 34, 2011
|
|
3ARB
 
 | | Ternary crystal structure of the NKT TCR-CD1d-alpha-galactosylceramide analogue-OCH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wun, K.S, Rossjohn, J. | | Deposit date: | 2010-11-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells
Immunity, 34, 2011
|
|
3ARD
 
 | | Ternary crystal structure of the mouse NKT TCR-CD1d-3'deoxy-alpha-galactosylceramide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, Beta-2-microglobulin, ... | | Authors: | Wun, K.S, Rossjohn, J. | | Deposit date: | 2010-11-27 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells
Immunity, 34, 2011
|
|
3ARE
 
 | | Ternary crystal structure of the mouse NKT TCR-CD1d-4'deoxy-alpha-galactosylceramide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wun, K.S, Rossjohn, J. | | Deposit date: | 2010-11-27 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells
Immunity, 34, 2011
|
|
3ARF
 
 | | Ternary crystal structure of the mouse NKT TCR-CD1d-C20:2 | | Descriptor: | (11Z,14E)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]icosa-11,14-dienamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wun, K.S, Rossjohn, J. | | Deposit date: | 2010-11-27 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells
Immunity, 34, 2011
|
|
3BLS
 
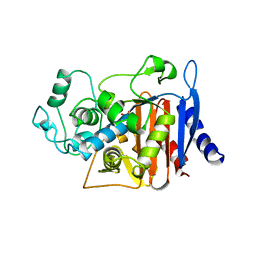 | | AMPC BETA-LACTAMASE FROM ESCHERICHIA COLI | | Descriptor: | AMPC BETA-LACTAMASE, M-AMINOPHENYLBORONIC ACID | | Authors: | Usher, K.C, Shoichet, B.K, Remington, S.J. | | Deposit date: | 1998-06-04 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of AmpC beta-lactamase from Escherichia coli bound to a transition-state analogue: possible implications for the oxyanion hypothesis and for inhibitor design.
Biochemistry, 37, 1998
|
|
3A40
 
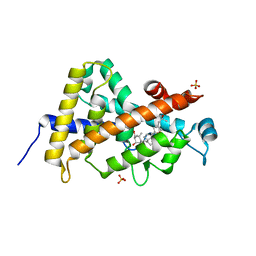 | | Crystal structure of the human VDR ligand binding domain bound to the synthetic agonist compound 2alpha-methyl-AMCR277B(C23R) | | Descriptor: | (1S,2S,3R,5Z,7E,14beta,17alpha,23R)-23-(2-hydroxy-2-methylpropyl)-2-methyl-20,24-epoxy-9,10-secochola-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Sato, Y, Antony, P, Huet, T, Sigueiro, R, Rochel, N, Moras, D, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2009-06-25 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-function relationships and crystal structures of the vitamin D receptor bound 2 alpha-methyl-(20S,23S)- and 2 alpha-methyl-(20S,23R)-epoxymethano-1 alpha,25-dihydroxyvitamin D3
J.Med.Chem., 53, 2010
|
|
3AXL
 
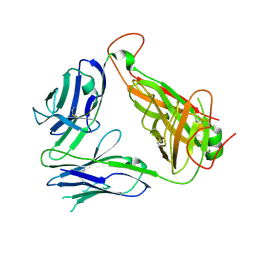 | | Murine Valpha 10 Vbeta 8.1 T-cell receptor | | Descriptor: | Valpha 10, Vbeta 8.1 | | Authors: | Patel, O, Rossjohn, J. | | Deposit date: | 2011-04-11 | | Release date: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A semi-invariant V(alpha)10(+) T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties
Nat.Immunol., 12, 2011
|
|
