7XER
 
 | | Structure of mTRPV2_Q525T | | Descriptor: | CHOLESTEROL, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEU
 
 | | Structure of mTRPV2_E2 | | Descriptor: | CHOLESTEROL, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEW
 
 | | Structure of mTRPV2_Q525F | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7WNM
 
 | | Structure of SARS-CoV-2 Gamma variant receptor-binding domain complexed with high affinity human ACE2 mutant (T27F,R273Q) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Ma, R.Y, Han, P.C, Wang, Q.H, Han, P. | | Deposit date: | 2022-01-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A binding-enhanced but enzymatic activity-eliminated human ACE2 efficiently neutralizes SARS-CoV-2 variants.
Signal Transduct Target Ther, 7, 2022
|
|
7XNF
 
 | | Structure of SARS-CoV-2 antibody P2C-1F11 with GX/P2V/2017 RBD | | Descriptor: | P2C-1F11 Heavy Chain, P2C-1F11 Lambda chain, Spike protein S1 | | Authors: | Jia, Y.F, Chai, Y, Wang, Q.H, Gao, G.F. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Cross-reaction of current available SARS-CoV-2 MAbs against the pangolin-origin coronavirus GX/P2V/2017.
Cell Rep, 41, 2022
|
|
7XSW
 
 | | Structure of SARS-CoV-2 antibody S309 with GX/P2V/2017 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Heavy Chain, S309 Lambda Chain, ... | | Authors: | Jia, Y.F, Chai, Y, Wang, Q.H, Gao, G.F. | | Deposit date: | 2022-05-15 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Cross-reaction of current available SARS-CoV-2 MAbs against the pangolin-origin coronavirus GX/P2V/2017.
Cell Rep, 41, 2022
|
|
7UKL
 
 | |
7WRI
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Han, P, Xie, Y, Qi, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-08 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
7WSK
 
 | | Crystal structure of SARS-CoV-2 Omicron spike receptor-binding domain in complex with civet ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Huang, B, Han, P, Qi, J. | | Deposit date: | 2022-01-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
7W88
 
 | |
7W82
 
 | | Crystal structure of maize RDR2 | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Du, X, Yang, Z, Du, J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of plant RNA-DEPENDENT RNA POLYMERASE 2, an enzyme involved in small interfering RNA production.
Plant Cell, 34, 2022
|
|
7W6R
 
 | | Structure of Bat coronavirus RaTG13 spike receptor-binding domain complexed with its receptor equine ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein, ... | | Authors: | Xu, Z.P, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2021-12-02 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding and structural basis of equine ACE2 to RBDs from SARS-CoV, SARS-CoV-2 and related coronaviruses
Nat Commun, 13, 2022
|
|
7W84
 
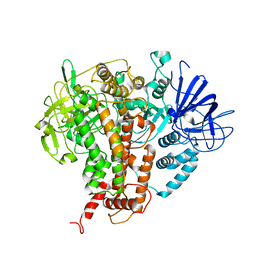 | |
7W6U
 
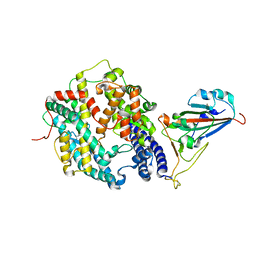 | | Structure of SARS-CoV-2 spike receptor-binding domain complexed with its receptor equine ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Xu, Z.P, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2021-12-02 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Binding and structural basis of equine ACE2 to RBDs from SARS-CoV, SARS-CoV-2 and related coronaviruses
Nat Commun, 13, 2022
|
|
7WXZ
 
 | | Crystal structure of the recombinant protein HR121 from the S2 protein of SARS-CoV-2 | | Descriptor: | Spike protein S2' | | Authors: | Zheng, Y.T, Ouyang, S, Pang, W, Lu, Y, Zhao, Y.B. | | Deposit date: | 2022-02-15 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A variant-proof SARS-CoV-2 vaccine targeting HR1 domain in S2 subunit of spike protein.
Cell Res., 32, 2022
|
|
7XBY
 
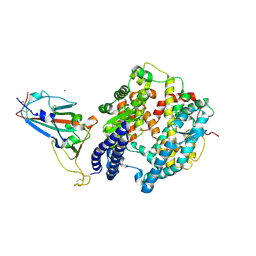 | | The crystal structure of SARS-CoV-2 Omicron BA.1 variant RBD in complex with equine ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, BROMIDE ION, ... | | Authors: | Xu, Z.P, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-22 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Binding and structural basis of equine ACE2 to RBDs from SARS-CoV, SARS-CoV-2 and related coronaviruses.
Nat Commun, 13, 2022
|
|
7XBG
 
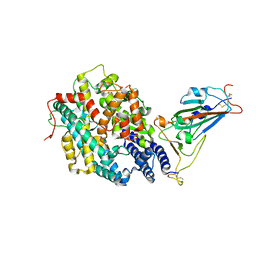 | | The crystal structure of RshSTT182/200 RBD-insert2-T346R-Y496G mutant in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7XBH
 
 | | The complex structure of RshSTT182/200 RBD bound to human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, RshSTT182/200 coronavirus receptor binding domain, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7XBF
 
 | | The complex structure of RshSTT182/200 RBD-insert2 bound to human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, RshSTT182/200 coronavirus receptor binding domain insert2, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7WRH
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Han, P, Xie, Y, Qi, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
7XAB
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 22d | | Descriptor: | 9-(cyclopropylmethoxy)-8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-5-(pyridin-4-ylmethoxy)pyrano[3,2-b]xanthen-6-one, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2022-03-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.00067449 Å) | | Cite: | Discovery of novel PDE4 inhibitors targeting the M-pocket from natural mangostanin with improved safety for the treatment of Inflammatory Bowel Diseases.
Eur.J.Med.Chem., 242, 2022
|
|
7XAA
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 21d | | Descriptor: | 8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-9-oxidanyl-5-(pyridin-4-ylmethoxy)pyrano[3,2-b]xanthen-6-one, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2022-03-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.100414 Å) | | Cite: | Discovery of novel PDE4 inhibitors targeting the M-pocket from natural mangostanin with improved safety for the treatment of Inflammatory Bowel Diseases.
Eur.J.Med.Chem., 242, 2022
|
|
