6CNW
 
 | | STRUCTURE OF HUMANIZED SINGLE DOMAIN ANTIBODY SD84 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, humanized antibody SD84h | | Authors: | Luo, J, Obmolova, O. | | Deposit date: | 2018-03-09 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin.
Science, 362, 2018
|
|
8SXP
 
 | |
6CNV
 
 | | INFLUENZA B/BRISBANE HEMAGGLUTININ FAB CR9115 SD84H COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR9114 Fab heavy chain, ... | | Authors: | Luo, J, Obmolova, G. | | Deposit date: | 2018-03-09 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin.
Science, 362, 2018
|
|
2KA5
 
 | | NMR Structure of the protein TM1081 | | Descriptor: | Putative anti-sigma factor antagonist TM_1081 | | Authors: | Serrano, P, Geralt, M, Mohanty, B, Pedrini, B, Horst, R, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2KLA
 
 | | NMR STRUCTURE OF A PUTATIVE DINITROGENASE (MJ0327) FROM METHANOCOCCUS JANNASCHII | | Descriptor: | Uncharacterized protein MJ0327 | | Authors: | Jaudzems, K, Mohanty, B, Geralt, M, Serrano, P, Wilson, I, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein NP_247299.1: comparison with the crystal structure.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2KL2
 
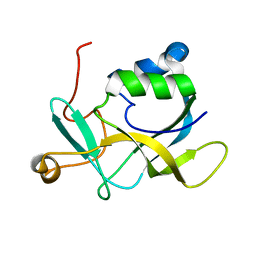 | | NMR solution structure of A2LD1 (gi:13879369) | | Descriptor: | AIG2-like domain-containing protein 1 | | Authors: | Pedrini, B, Serrano, P, Mohanty, B, Geralt, M, Herrmann, T, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
8U52
 
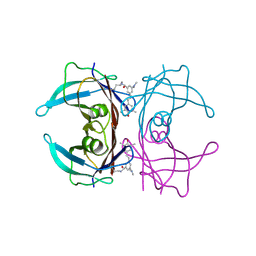 | | Co-crystal structure of human transthyretin conjugated to the stilbene substructure derived from reaction with the fluorogenic covalent kinetic stabilizer A2 | | Descriptor: | S-(4-fluorophenyl) 3-(dimethylamino)-5-[(E)-2-(4-hydroxy-3,5-dimethylphenyl)ethenyl]benzenecarbothioate, Transthyretin | | Authors: | Yan, N.L, Nugroho, K, Kline, G.M, Basanta, B, Lander, G.C, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2023-09-11 | | Release date: | 2025-03-05 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The conformational landscape of human transthyretin revealed by cryo-EM.
Nat.Struct.Mol.Biol., 32, 2025
|
|
4KTD
 
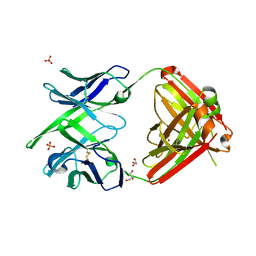 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE136, from non-human primate | | Descriptor: | GE136 Heavy Chain Fab, GE136 Light Chain Fab, GLYCEROL, ... | | Authors: | Poulsen, C, Tran, K, Standfield, R, Wyatt, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3O0F
 
 | |
3PAY
 
 | |
3NL9
 
 | |
3PL0
 
 | |
2YPI
 
 | |
3OZ2
 
 | |
3OHG
 
 | |
3OYV
 
 | |
3NPD
 
 | |
3N8U
 
 | |
3PXP
 
 | |
3RJ0
 
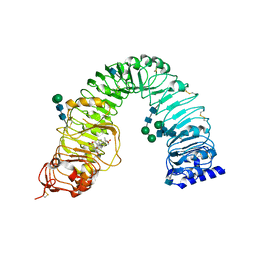 | | Plant steroid receptor BRI1 ectodomain in complex with brassinolide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinolide, Protein BRASSINOSTEROID INSENSITIVE 1, ... | | Authors: | Hothorn, M. | | Deposit date: | 2011-04-14 | | Release date: | 2011-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Structural basis of steroid hormone perception by the receptor kinase BRI1.
Nature, 474, 2011
|
|
3R4R
 
 | |
1FO0
 
 | | MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX | | Descriptor: | NATURALLY PROCESSED OCTAPEPTIDE PBM1, PROTEIN (ALLOGENEIC H-2KB MHC CLASS I MOLECULE), PROTEIN (BETA-2 MICROGLOBULIN), ... | | Authors: | Reiser, J.B, Darnault, C, Guimezanes, A, Gregoire, C, Mosser, T, Schmitt-Verhulst, A.-M, Fontecilla-Camps, J.C, Malissen, B, Housset, D, Mazza, G. | | Deposit date: | 2000-08-24 | | Release date: | 2000-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a T cell receptor bound to an allogeneic MHC molecule.
Nat.Immunol., 1, 2000
|
|
8SIT
 
 | |
8SIS
 
 | |
8SIR
 
 | |
